Abstract
The success of deep learning over the traditional machine learning techniques in handling artificial intelligence application tasks such as image processing, computer vision, object detection, speech recognition, medical imaging and so on, has made deep learning the buzz word that dominates Artificial Intelligence applications. From the last decade, the applications of deep learning in physiological signals such as electrocardiogram (ECG) have attracted a good number of research. However, previous surveys have not been able to provide a systematic comprehensive review including biometric ECG based systems of the applications of deep learning in ECG with respect to domain of applications. To address this gap, we conducted a systematic literature review on the applications of deep learning in ECG including biometric ECG based systems. The study analyzed systematically, 150 primary studies with evidence of the application of deep learning in ECG. The study shows that the applications of deep learning in ECG have been applied in different domains. We presented a new taxonomy of the domains of application of the deep learning in ECG. The paper also presented discussions on biometric ECG based systems and meta-data analysis of the studies based on the domain, area, task, deep learning models, dataset sources and preprocessing methods. Challenges and potential research opportunities were highlighted to enable novel research. We believe that this study will be useful to both new researchers and expert researchers who are seeking to add knowledge to the already existing body of knowledge in ECG signal processing using deep learning algorithm.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
The applications of Machine Learning (ML) techniques in medical biosignals data and sensors have received a tremendous attention from researchers in the last decades. However, the advent of Internet of medical Things (IoMTs), such as implantable medical device and personal wearable devices led to the large volume and varieties of data called big data (Belle et al. 2015; Corradi et al. 2019; Konan and Patel 2018). This made the use of traditional ML and feature engineering not suitable to process these data. More so, ML techniques depend largely on hand-crafted features which require deep domain knowledge to extract effective features and hence, time consuming (Hammad et al. 2019; Pyakillya et al. 2017; Taherisadr et al. 2018). Moreover, ML models suffer a great deal of problem of vanishing gradient and over fitting which affects the performances of the training models(Z.-J. Yao et al. 2018).
In respond to the challenges facing the shallow ML techniques as outlined in the previous section, Deep Learning (DL) algorithms emerges to solve the challenges of the shallow ML techniques. The DL algorithms perform better as the dataset becomes larger and the problem of vanishing gradient is handled through the application of the DL. The DL can extract information of clinical relevance hidden in large volume of health care data. Consequently, help in treatment, decision making and prevention of health conditions (Alom et al. 2019). The DL as a sub-class of ML has become a buzz word that dominates Artificial Intelligence (AI) applications and researches in recent years.
The popularity attracted by the DL is as a result of the successes recorded in handling complex data analytic problems such as, computer vision (Voulodimos et al. 2018), object recognition, detection, and segmentation (Wang 2016), face recognition(Shepley 2019), speech recognition (Nassif et al. 2019), image classification and localization(Pak and Kim 2017), natural language processing (Young et al. 2018), robotics (Pierson and Gashler 2017), medical imaging (Lee et al. 2017) and so on, better than the traditional ML techniques and feature engineering (Ince et al. 2017; Kwon et al. 2020; Tobore et al. 2019; Voulodimos et al. 2018). Another major difference between DL and traditional ML is in the presentation of data during model training (Miotto et al. 2018) and the way features are extracted (Alom et al. 2019). The early years of 21st century marked the “age of deep learning” (Voulodimos et al. 2018).
However, DL rose to become popular starting from the last decade, when Krizhevsky et al. made a high performance improvement on their convolutional neural networks model (AlexNet), that reduces top-5 error by 10% in 2012 and became the best model in the 2012 ImageNet Large Scale Visual Recognition Competition (ILSVRC-2012) (Krizhevsky et al. 2012). Other reasons associated with the rise of DL include the large availability of data and advances in computing power. The DL generalizes better when model with huge amount of data during model training. This would lead to computational cost which is handled by Graphic Processing Units (GPU), a special kind of processors that allow massive parallel computing. More so, the developments of software frameworks such as Keras, TensorFlow, Teano and PyTorch (Voulodimos et al. 2018), have made researchers to focus on the DL structures. Another common reason is the fact that the DL do not need hand-crafted features or feature engineering processes to extract features for training, instead, they can perform feature extraction, feature selection and classification automatically (Li et al. 2020). The DL are considered the promising technology suitable for big data generated from the medical applications. The DL models have also proven to produce state-of-the-art performances and more domains are gravitating towards it.
Electrocardiogram (ECG) is one of the most successful heart diseases diagnostic tools that have attracted a tremendous attention from researchers for decades. The ECG signals are biosignals that can help to detect heartbeats irregularity through calculating the bioelectrical and muscle activity of the heart (Shankar and Babu 2020). The Biosignals are electrical, thermal, mechanical, or other signals measured over the time from the human body (Ganapathy et al. 2018). The ECG instrument keeps records of the heartbeat’s activity continuously over time, which can be used for diagnosis. It records the electrical activities of the heart by placing the noninvasive electrodes (leads) on the subject’s body, typically, the chest and limb (Assodiky et al. 2017; Isin and Ozdalili 2017; Wang et al. 2019). These leads measure the voltage variations triggered by an involuntary impulses of the cardiac cells as they make the heart contract. In effect, these variations form the heartbeats that are seen in the form of series of waves. Time information of the electrical activity can be traced from the morphologies of the waves (Abdeldayem and Bourlai 2019) and used to detect heart and coronary related pathologies. The application of DL in ECG signals for various medical and health care applications has gained tremendous attention by the research community in the last decade. The use of these applications is essentially important to automatically diagnose patients’ cardiovascular diseases where there is a shortage of experienced and qualified medical doctors to interpret the ECG signals or even compliment the function of Cardiologist (Ribeiro et al. 2020).
This review is focused on related studies that use DL in ECG signals for various analyses from different domains.
Figure 1 illustrates the Google trends for DL compared with ECG. The search was performed using web search, for all categories, worldwide and from January, 2010 to May, 2020Footnote 1. It can be observed that the trend in the graph (a) shows interest vs. time on a scale out of 100, where the score 0 means that the sufficient data is not present for the term. The score of a 100 signifies that the term is in its peak and more popular than any other topic at that time. Figure 1 (a) shows the steady rise of interest in both ECG and DL, with ECG reaching its peak value of 100 in February, 2020. The DL on the other hand, had minimal interest from 2010 to 2012, but shows a growing interest after 2012 onward. This may be as a result of the renewed interest and popularity gained by a DL model when CNN-based model (AlexNet) reduced top-5 error by 10% in 2012 and won the yearly ILSVRC-2012 competition (Krizhevsky et al. 2012). Figure 1(b) shows the top 5 popular countries where the search terms were more popular, sorted by interest for “deep learning + deep neural networks”; with China, South Korea, Japan, Germany and Taiwan on the top list.
There are a number of review papers found in the literature on the applications of DL in ECG signals from different perspectives Bote-Curiel et al. 2019; Faust et al. 2018; Ganapathy et al. 2018; Hong et al. 2020; Rim et al. 2020; Tobore et al. 2019; Z.-J. Yao et al. 2018). Most of these papers mainly work on the applications of DL in physiological signals analysis as a whole including ECG and for various medical (and/or healthcare) applications. However, evidence from the literature reveals the applications of DL in ECG-based biometric systems for human identification Bajare and Ingale 2019; Byeon et al. 2020; P.-L. Hong et al. 2019) and authentication (Hammad et al. 2018, 2019; Hammad and Wang 2019). Also, there are applications of DL in ECG-based driver drowsiness detection (Abbas 2020), stress level classification (Rastgoo et al. 2019) and pilot load prediction towards mitigating the risks of accidents (Xi et al. 2019). Nevertheless, (Hong et al. 2020) conducted a systematic review and focused on applications of DL in ECG data, considering the model architecture, source of data and the tasks perspectives, but it was not sufficiently extensive.
This study improves on (Hong et al. 2020) to presents a comprehensive systematic review and meta-data analysis on the applications of DL in ECG signals with respect to different domains that were not covered in the previous survey.
The subsequent sections of this study are structured as follows: Fig. 2 shows the general organization of the complete review. In Sect. 2, we present evolution of DL. Section 3 discusses the overview of ECG. In Sect. 4, we review related works. Section 5 outlines the systematic literature review processes. In Sect. 6, the paper presents the general discussion and meta-data analysis. Section 7 discusses challenges and future research directions while conclusion is presented in Sect. 8.
The summary of the contribution of this study are as follows: The study present taxonomy of domains for DL in ECG-based applications: medical/healthcare, biometric/security and driving. The paper highlights unresolved challenges and pointed out future directions. To the best knowledge of the authors, this is the first review to study the applications of DL in ECG signals with respect to different domains. We carried out meta-data analysis of the DL approaches in ECG signals based on their application domains, ECG preprocessing methods, application area, DL application tasks, DL models, DL model performances, datasets sources, and the architectures.
2 The evolution of Deep Learning
The DL is a new generation of Artificial Neural Networks (ANNs), a subset of ML a subset of AI (Alom et al. 2019; Nguyen et al. 2019). The emergence of AI can be traced back to the 1950s, when scientists thought computers could do things at the level of human intelligence. Notably, in 1950, Alan Turing asked a question, “Can machines think?” This question led to the journey of inventions of models from knowledge-based systems (also called symbolic AI) to ML models (Chollet 2018; Mohammed et al. 2016). In the 1956, John McCarthy et al. from IBM and Claude Shannon organized the first AI conference at Dartmouth College, USA, where the term “Artificial Intelligence” was first coined and used later in the second conference. The AI is the simulation of human intelligence on computers to mimic human intelligence (Jiang et al. 2017; Nguyen et al. 2019). This involves making computers to perform tasks like or even more than how a human would/could. Therefore, AI deals with the automation of human intelligence that would make machines perform tasks intelligently as humans (Chollet 2018).
The ML paradigm came in handy to curtail the limitations of the symbolic AI, that could not handle explicit rules for computing more complex and fuzzy problems like the speech recognition, computer vision, image processing, text classification, natural languages processing and pattern recognition. Instead of following some predefined set of rules, mapping input data to output data as it is with the symbolic AI, ML system is “trained” to learn (just like humans learn by experience), by supplying input data with labeled answers into the system. The ML enables computers to learn without programming explicitly (Chollet 2018; Mohammed et al. 2016). Learning refers to a procedure consisting of tuning the model parameters such that the learned model can perform a specific task (Alom et al. 2019). This enables an AI system to sieve information from raw data and present inferences based on input-output relationships thereby learning from experience for better generalization of new raw data (McBee et al. 2018). The ML algorithms that have gained popularity includes support vector machine (SVM), k-nearest neighbor (KNN), decision trees (DT), multilayer perceptron (MLP) and so on. The ML therefore provides a glimpse of hope to achieve the ultimate goal of AI – the explicit automation of human intelligence. Figure 3 shows the relationship between AI, ML, ANN and DL.
When it comes to performing work, intelligence is the central difference between humans and machines. Humans can learn using experience to take decisions, but machines cannot, they are built to perform specific and predefined set of tasks. The ML aims at bridging this gap. The advancement made in bridging this gap is further made possible with the introduction of ANN.
An ANN is a software simulation of how the biological brain functions, an extremely simplified model of the animal brain. The ANNs have witnessed three developmental waves so far from the literature. It started with the introduction of perceptron in 1950s, followed by the development of perceptron with backpropagation in 1970s and the introduction of DL in 1990s. The introduction of the first ANN model was by a neurophysiologist, Waren McCulloch and a mathematician, Walter Pitts, in 1943. Since then, scientists made contributions with Frank Rosenblatt who invented the first perceptron in 1957 (Mohammed et al. 2016). Perceptron is the simplest representation of biological neuron in the ANN.
Figure 4 shows a typical representation of a perceptron (the right picture) and a biological neuron (the left picture). The inputs (x1, x2,x3, …,xn) represent the dendrites carrying inputs data. These data inputs are multiplied with randomly generated weights(w1, w2,w3,…, wn) for each input data. The dot matrix product of (x1, x2,x3,…, xn) and (w1, w2,w3,…,wn) are summed up and a pre-determined value also called bias is added, this represent the body of a biological neuron. The activation, f, which represents the axon, is computed (using what is called step function). But this function can only estimate linear relations in the data. However, recent activation functions, such as sigmoid, Rectified Linear Unit (ReLU) and hyperbolic tangent (tanh) allow estimating complex and nonlinear relations in the input data and provide a normalization effect on the output data. The output, y, outputs one (1) if the resultant is more than some certain threshold value and the output is zero (0), if otherwise. The output y is computed using Eq. (1).
In 1969, a book published by Marvin Minsky and Seymour Papert, called “Perceptrons” pointed the limitations of perceptron, which could not able to solve more complex features like XOR logic, and not able to tackle non-linearity features of input data in ANN. The authors of the book argued that the single perceptron approach to ANN could not be translated effectively into multi-layered ANN. As a result, ANN projects suffered from funding by organizations. However, in 1981, Paul Werbos proposed the first efficient ANN with backpropagation. Backpropagation algorithm works by fine-tuning the weights of an ANN based on the error rate computed in the previous iteration in order to reduce the difference between the actual output and the desired output (error), thus increasing its generalization ability (Fig. 5). In 1986, Rumelhart et al. work propagated the use of backpropagation and introduced hidden layers (Mohammed et al. 2016; Schmidhuber 2015). Figure 5 depicts a simple representation of an ANN with backpropagation. It may consist of tens to hundreds of neurons arranged in layers, with each layer connected to the layers on both sides. It has three parts consisting of the input unit (left), the hidden layer(s) (middle) and the output layer (right) that generates the results.
Figure 5 represents the working principle of backpropagation, the steps include first, input data, X is supplied into the network (1). A set of randomly generated weights, W, are multiplied for each X and summed with a bias value (2). The output layer (3) computes the output of the trained model. The loss function is calculated (4) and a backpropagation technique is applied (5) to backtrack to the hidden layers to fine-tune the weights such that the loss function is reduced. This process continues until the model is well trained or the number of epochs is reached.
The DL is based on the ANN that apply linear and nonlinear transformations of the input data from the input layer through the multiple hidden layers of processing units to the output layer (Ganapathy et al. 2018). These multiple processing layers imitate the human brain in how they represent data with multiple levels of abstraction. This helps the network to understand multimodal information, thereby implicitly capturing salient structures of large-volume of data (Voulodimos et al. 2018). Hinton et al. published a paper in 2006 (Hinton et al. 2006) attributed to be the first to come up with the concept and method of the DL. DL is a form of ML that enables machines to learn from experience and successive of concepts of the world (Kim 2016). Conversely, the form of ML that makes machines learn using only three layers of representations (input, hidden and output layers) of the data is sometimes called “shallow” learning or shallow model (Z.-J. Yao et al. 2018). The term “deep” in DL connotes the idea of hierarchical layers of representations, with modern DL having tens or hundreds hierarchical layers of representations (Chollet 2018; Faust, Hagiwara, et al., 2018). The major difference between DL and traditional ANNs is in the number of hidden layers (Fig. 6), their connections and the capability to learn meaningful abstractions of the input data (Miotto et al. 2018; Tobore et al. 2019). However, the performance largely dependent on the nature of the data representation presented to the model (Kim 2016). From 1990s, DL has successfully been used in different applications, it grows to become probably the most well-known field of AI (Bote-Curiel et al. 2019). For a more detailed timeline history and evolution of DL, the reader is referred to (Emmert-Streib et al. 2020) as the detail is not within the scope of the work. The DL is sometimes called the universal learning approach because it has been found applicable in almost all areas. Therefore, DL is task-independent (Alom et al. 2019). However, the potentials and opportunities of DL architectures are still being explored.
Figure 6 depicts the comparison between ANNs and DL. The ANNs usually have three layers where learning takes place towards the output. The DL consists of many hidden layers, tens to hundreds of layers where DL learn and fine-tune errors using backpropagation algorithm to extract salient features and structures of inputs for a more generalizable models. Learning approaches in ML and DL are classified into supervised learning, unsupervised learning, semi-supervised learning and reinforcement learning (Alom et al. 2019; Mohammed et al. 2016).
2.1 Deep learning architectures
In this section, we discuss the DL architecture commonly used in ECG signal processing.
2.1.1 Deep neural networks
The Deep Neural Networks (DNNs) are special types of ML techniques with deeper networks than the traditional network. Even though DL can be referred to as DNNs generally, but for the sake of clarity in this review, DNN is considered as the conventional ANN with at least two hidden layers. Hence, DNN is a particular ANN with at least 4 layers, comprises of input, hidden layers and output layer (Sannino and De Pietro 2018). The use of DNNs suffers from the vanishing gradient problem; moreover, it is difficult to train. Over the years, many researchers tried to solve these challenges which led to the introduction of different kinds of DL architectures (Z.-J. Yao et al. 2018). Figure 7 shows the structure of the DNN.
2.1.2 Convolutional neural networks
Convolutional Neural Networks (CNNs), based on the human visual cortex was first proposed in 1962 by Hubel and Wiesel (Hubel and Wiesel 1962) and is the most utilized neural network for computer vision and video recognition (Shrestha and Mahmood 2019). It is a DL-based technique designed to automatically and adaptively learn features from an input image/data and classifies the input data into the desired classes. The CNN structure has three building blocks; the convolution layers, the pooling layers and the fully connected layers. The convolution and pooling layers are normally employed to extract features and fully connected layer is used for classification (Voulodimos et al. 2018; Yamashita et al. 2018). The concept of CNN was first proposed by Fukushima(Fukushima, 1995), but it was not widely used until when Lecum et al. in 1998, designed CNN for document analysis and recorded a good result for handwritten digit classification(LeCun et al. 1998). Then CNN rose to become popular about 14 years later when Krizhevsky et al. made a high performance improvement on their model known as AlexNet, which won the yearly ILSVRC-2012 (Krizhevsky et al. 2012). The year after, in 2013, ZF Net was developed which was an enhancement on AlexNet (Zeiler and Fergus 2014). It was followed by the development of GoogLeNet, a 22 layers deep network by researchers from Google Inc.(Szegedy et al. 2015).
The GoogLeNet won the ILSVRC 2014 challenge. Simonyan and Zisserman (Simonyan and Zisserman 2014), proposed VGGNet (also called VGG). They won the first and the second places in the ILSVRC 2014 for localization and classification tasks respectively. The ResNet, another powerful CNN architecture was proposed by He et al. (He et al. 2016) referred to as residual learning framework. The model won the first place on the ILSVRC 2015 classification task. There are other variants of the ResNet such as the ResNet50, ResNet34 and ResNeXt architectures. SqueezeNet (Iandola et al. 2016) is a model based on ResNet with 510 times less memory requirements and this is achieved through deep compression technique. Densely connected CNN (DenseNet) (Huang et al. 2017) proposed based on the deep CNN model. A DenseNet was designed in such a way that every layer is connected to every other layer in a feed forward network, it gives it an edge over the previous CNNs, such that it reduces the vanishing-gradient problem, strengthen feature propagation, encourage feature reuse, and substantially reduce the number of parameters required. The CNN is considered the most used DL for classification (Pourbabaee et al. 2018; Schmidhuber 2015; Shi et al. 2020). The CNN has been applied in computer vision (Khan et al. 2018; Voulodimos et al. 2018), language translation (Yin et al. 2017), image segmentation (Kayalibay et al. 2017), object recognition (Kulik and Shtanko 2020) and so on. The architecture of the CNN (see Fig. 8) shows the different components of the architecture.
The architecture of CNN (Sengupta et al. 2020)
An input data e.g. an image (an m × m × r), where m is the height and width of the image and r is the number of channels) from the input layer passes through the convolution layer and pooling layer. The features of the input image also called input tensor, are extracted and classification of this image takes place at the fully connected layers where Softmax function is applied to classify an object with probabilistic values [0, 1](Sengupta et al. 2020).
The convolutional layer: This is the first layer that receives the input data. The Kernel, k, an n × n × q, where n is smaller than the dimension of the input image, m, and q can be the same as r, which is an array of numbers, called a tensor. The dimension of k can be m - n + 1. A matrix multiplication between each element of k and the input tensor is computed at each location of the tensor while it hovers over the input tensor at a stride length (distance between two successive kernels) and summed to obtain the output value in the corresponding position of the output tensor, called the feature map. The tensor hops using the same stride value and repeats the process until the entire image is traversed (see Fig. 9). The convolution operation can be performed over multiple convolutional layers thereby capturing high-level features for better performances. The key feature of a convolution operation is weight sharing: kernels are shared across all the image positions. In some cases where the kernels doesn’t fit perfectly on the input, valid padding or zero padding is applied. The former is when convolved feature is reduced in dimensionality as compared to the input. Valid padding drops the part where the kernel does not fit. Zero padding pads with zeros so that the filter fits in. In the convolution operation, the size of the kernels which is typically 3 × 3, the number of kernels, padding and stride value are decided prior to the training. These parameters are maintained to pooling layer. The convolution operation is mathematically expressed as in Eq. (2) (Alom et al. 2019):
Where.
\({\text{x}}_{\text{j}}^{\text{l}}\)= current layer output
\({\text{x}}_{\text{i}}^{\text{l-1}}\)= previous layer output
\({\text{k}}_{\text{ij }}^{\text{l}}\)= current layer kernel
\({\text{b}}_{\text{j}}^{\text{l}}\)= bias of the current layer
\({M}_{j}\)= selection of the input maps
The non-linear operation which is the use of activation function applied on the output of the convolution operation. Typically, the sigmoid function is used but the most common nonlinear activation function used presently is the rectified linear unit (ReLU) that simply computes the function: f(x) = max (0, x).
Illustrates convolution operation with a stride of 1, a kernel size of 3 × 3 and no padding (Yamashita et al. 2018)
The pooling layer
This layer basically performs sub-sampling operation on the convolved features (feature map) for the next convolutional layer. This is with the aim to decrease the computational power required to process the data by dimensionality reduction and help reduce the chance for overfitting. However, it retains important information. There are different pooling operations such as max, average and sum pooling. Max and average pooling are the most popular (Alom et al. 2019; Voulodimos et al. 2018). The sub-sampling operation is computed using Eq. (3).
,
The fully connected (FC) layer
This layer flattens the output feature maps of the convolutional or pooling layer from the 2-dimensional (2D) feature maps into 1-dimensional (1D) array of numbers (or vector) and feed it into a fully connected layer for classification. Every input is connected to every output by a learnable weight. Each fully connected layer is followed by a nonlinear function, such as ReLU. Finally, an activation function such as softmax or sigmoid is used to classify the input image(Yamashita et al. 2018).
2.1.3 Deep recurrent neural networks
Deep recurrent neural network is a variant of the recurrent neural networks (RNNs) with at least two hidden layers, where the current output depends on the computations performed previously, hence the name recurrent (Andersen et al. 2019). A RNN carries out training by remembering the previous step from the current step. This is different from feed forward neural network that trains only in the forward direction. This makes it inefficient for sequential data with dependencies like the time series predictions, speech recognition, voice semantic recognition etc. A RNN is said to possess a “memory” kind of concept that remembers the previous computation done before the current state, giving the RNN a kind of contextual information. Another important feature of the RNN is that, it provides same parameters for input layer through the hidden layers to output layer. Thereby, reduce the complexity of parameters in contrast with other ANN. The training of a RNN is preferably performed with data that have interdependencies to maintain information about what occurred in the previous interval (Tobore et al. 2019). However, the advantages of the RNN came with a disadvantage of vanishing gradient problem (Bengio et al. 1994; Shrestha and Mahmood 2019). To this effect, long short-term memory unit (LSTM) was proposed which effectively handle this challenge (Hochreiter and Schmidhuber 1997). Another RNN-based architecture is gated recurrent unit (GRU) which is a special case of LSTM. It has equivalent performance with LSTM but faster than the LSTM (Lynn et al. 2019; Wang et al. 2018). In this review, we only present the architecture of LSMT as the most used variant of RNN (Yildirim 2018). The architecture of LSTM is depicted in Fig. 10.
Architecture of LSTM (Tobore et al. 2019)
The LSTM extends the memory retention capability of RNN that retains it is value as a function of it is inputs data because RNN suffers from lags. The LSTM structure has three gates (input, forget and output gate) that direct the flow of information from previous to current state to the memory cell and to the next memory cell (Sharma et al. 2020). The input gate is establishes when the flow of the new information into the memory should take place. The forget gate controls how long the stored information should be retained giving free space for new data. The output gate decides when the stored information in the cell is used in the output (Hernández-Blanco et al., 2019). Figure 10 depicts the LSTM representation where, It, Ot, and Mt are the current input, output and memory state of the LSTM cell. It−2 and It−1, Ot−2 and Ot−1, Mt−2 and Mt−1 are the inputs, outputs, and memory respectively for previous time steps. Ot+1, and Mt+1 represent subsequent output and memory state respectively for subsequent time step input It+1. I, O, M represent the recurrent input, output and memory state respectively for a simplified LSTM cell operation and Wr is the weight for the computation in the cell. LSTM temporal information processing ability makes it popular and widely used (Alom et al. 2019). The memory cell \({c}_{t}\) is updated, while at same time produces output vector \({h}_{t}\) based on the following equations (Antczak 2018):
Where.
\({x}_{t}\)= input vector
\({f}_{t}\), \({i}_{t} and {?}_{t}\) = activation vectors of input, forget and output gates
\({\sigma }_{g}\), \({s}_{c} and {s}_{h}\) = activation functions, typically \({\sigma }_{g}\) is the sigmoid function, \({s}_{c} and {s}_{h}\) are tanh
\(\circ\)= denotes an operator called Hadamard product
2.1.4 Restricted Boltzmann Machines
The Boltzmann machines (BMs) are a kind of a bidirectional connected Networks with symmetrically coupled stochastic visible and hidden units. The visible units represent the first layer of the networks and correspond to the components of an observation whereas the hidden units model the dependences between these components of observations. Each of these units updates it is state over time in a probabilistic manner depending on the states of the neighboring units. This makes the learning of BMs computationally intensive. Restricted BM (RBM) is a parameterized generative stochastic ANN with undirected interactions between pairs of visible and hidden units. The RBM was designed to impose restrictions on the network topology in BM, thereby reducing the learning complexity of its parameters (Fischer and Igel 2012, 2014; Upadhya and Sastry 2019). Therefore, RBM becomes a variation to BM by given a restriction in the intra-layer connection between the visible and hidden units, forming a bipartite graph structure, hence the named RBM (Sengupta et al. 2020). Paul Smolensky was the first to introduce the RBM in 1986. However, he called it with the name Harmonium (Smolensky 1986).
The RBM has the ability to learn the input probability distribution in supervised as well as unsupervised approach, hence, it is popularity as a DL framework (Sengupta et al. 2020). There are two main DL architectures that incorporates RBM as learning module, namely, Deep Belief Networks (DBN) and Deep Boltzmann Machines (DBM) both are considered to belong to the “Boltzmann family” (Voulodimos et al. 2018). Special emphasis is given to DBN in this study. DBN is a model that is based on the combination of two different types of ANN. Specifically; DBNs amalgamate RBMs as the input unit with Deep Feedforward Neural Networks (D-FFNN) which forms the output unit (Emmert-Streib et al. 2020). Figure 11 depicts the structure of the DBN.
The DBN has undirected connections between it is top 2 layers and directed connections between all its subsequent layers (Voulodimos et al. 2018) (see Fig. 11). The DBN is initialized by greedy layer-wise training of the RBM Tobore et al. 2019; Voulodimos et al. 2018; Z.-J. Yao et al. 2018). Hinton and Salakhutdinov (Hinton and Salakhutdinov 2006) first proposed DBN and introduced the greedy layer-by-layer unsupervised learning algorithm that allows efficient training of these deep, hierarchical models in DBN. The DBN as a generative graphical model learns to extract a deep hierarchical representation of the training data. They model the joint distribution between observed vector x and the hidden layer as in Eq. (9).
Where \(\varvec{x}={\varvec{h}}^{0}, \varvec{p}\left({\varvec{h}}^{\varvec{k}}|{\varvec{h}}^{\varvec{k}+1}\right),\) is a conditional distribution for the visible units at level \(\varvec{k}\) conditioned on the hidden units of the RBM at level\({ \varvec{k}+1}^{ }\)and \(\varvec{p}\left({\varvec{h}}^{\varvec{l}-1}|{\varvec{h}}^{\varvec{l}}\right)\) is the visible-hidden joint distribution in the top-level RBM (Voulodimos et al. 2018).
2.1.5 Autoencoders
An Autoencoder (AE) is an unsupervised DL approach originally proposed by LeCun et al., in 1987 (LeCun et al. 1998). It involves dimension reduction of the input data and reconstruction of the input in the output layer (Shrestha and Mahmood 2019). An AE is a network of three layers; it becomes a deep AE with multiple hidden layers. Both the input layer and output layer have the same number of units, represented with the same dimensionality and the hidden layers typically have fewer units that encodes the inputs in a more compressed form (Sengupta et al. 2020; Tobore et al. 2019). The AE architecture is presented in Fig. 12. Training of AE involves two phases: The encoder and the decoder. The network is trained using backpropagation. During the encoding phase, the inputs are encoded into some hidden representations using the weight metrics of the lower half layer, and in the decoding phase, it tries to reconstruct the same input from the encoding representation using the metrics of the upper half layer. The encoding and decoding phases can be mathematically expressed as in Eqs. (10) and (11) respectively.
Where \({\varvec{x} }^{ }\)and \({\varvec{x}}^{\varvec{\text{?}}}\)represents the input vector and reconstructed input vector in the output layer respectively. Variable \(\varvec{w}\) and \(\varvec{b}\) are the parameters to be turned, \({\varvec{w}}^{\varvec{\text{?}}}\) and \(\varvec{c}\) is the transpose of \(\varvec{w}\), and the bias of the output layer respectively; \({\varvec{y}}^{ }\)is the hidden representation and \(\varvec{f}\) is the activation function. The parameters are updated using the following Equations:
Where \({\varvec{w}}_{\varvec{n}\varvec{e}\varvec{w}}\) and \({\varvec{b}}_{\varvec{n}\varvec{e}\varvec{w}}\) stands for the updated parameters of \(\varvec{w}\) and \(\varvec{b}\) respectively and \(\varvec{E}\) is the reconstructed error of input at the output layer (Sengupta et al. 2020).
Architecture of Autoencoder (Sengupta et al. 2020)
2.1.6 Generative adversarial networks
Generative Adversarial Network (GAN) is a DL architecture with unsupervised learning approach proposed by Goodfellow et al. in 2014 (Goodfellow et al. 2014). The GAN have two networks; generative and discriminator networks, that compete against each other in a zero-sum game simultaneously (Alom et al. 2019). The generative model tries to capture the data distribution whereas the discriminative model learns to estimate the probability of a sample either coming from training data or the distribution captured by the generative model. This can be viewed as a minmax two player game between these two models as the generative models produce adversarial examples while discriminative model trying to identify them correctly and both try to improve their efficiency until the adversarial examples are indistinguishable from the original ones (Sengupta et al. 2020). Figure 13 illustrates the flow of information in GAN deep network.
For a comprehensive discussion about the comparison between CNN and RNN, DBM and DBN, AE and RBM architectures, the reader is referred to (Tobore et al., 2019) as it is beyond the scope of this work.
3 The synopsis of Electrocardiogram
An electrocardiogram, usually abbreviated as “ECG” or “EKG” is a form of a test that provides the measurement of electrical signals generated from the heartbeat activity. It shows the condition of the heart and tells the status of various cardiovascular diseases (CVDs). ECG is a non-invasive and non-expensive tool, efficient in diagnosing cardiac disorders such as arrhythmia, by continuous monitoring of the ECG. The signals give information that can aid in analyzing and understanding the cardiac activity of a person such as heart rate, rhythm and morphology (Al Rahhal et al. 2016; Apandi et al. 2018; Park et al. 2019). Typically, the information provided by ECG test are the information of how long it takes for the electrical wave to pass through the heart by measuring time intervals on the ECG. This can help doctors to determine if the electrical signal passing through the heart is normal or slow, fasts or irregular. Secondly, measuring the amount of electrical wave passing through the heart muscle would help a cardiologist to diagnose if a part of the heart is overworked or too large. Electrocardiography is over a century old method with an established role in the care of patients with documented or suspected CVDs (Ribeiro et al. 2019). Other biosignals studied in the literature include Electromyography (EMG), Electroencephalography (EEG), Electrooculography (EOG), Photoplethysmography (PPG), phonocardiography (PCG), blood pressure and so on. The ECG is one of the biosignals or physiological signals, others includes EMG (concerns about changes to skeleton muscles), EEG (concerns about changes to the brain measured from the scalp), EOG (which concerns about changes to corneo-retinal potential between the front and the back of the human eye) (Rim et al. 2020), PPG (regards the volumetric changes of an organ over the time by recording changes in light absorption), blood pressure, and so on.
The ECG happened to be the first test tool developed in 1895 by Willem Einthoven and was found applicable in medical diagnosis (Ganapathy et al. 2018). The ECG rose to become one of the most widely used test tool for the CVDs disorders (Mincholé et al. 2019). Other areas where ECG is found applicable are: ECG-based biometric systems that have been proposed (both single and multimodal methods) for human identification and authentication using ECG as the physiological trait. In addition, ECG-based detection of driver drowsiness and stress level systems have been proposed in the literature to aid in reducing the rate of accidents. The ECG has also been used to predict the size and location of the heart as well as to locate the wound in the heart, and to ascertain the effectiveness of a drug (Byeon et al. 2020).
3.1 Types of ECG Machine
There are different types of ECG suitable for different situations and conditions. They can be used to measure the ECG signals either using in-the-person, on-the-person or off-the-person methods. The following are some of the commonly used types of the ECG machines:
-
The resting 12-leads ECG machine: This is considered the standard ECG for measuring heartbeat. With the 12 leads, more comprehensive signals can be obtained during the ECG measurement (Ribeiro et al. 2020). As demonstrated in (Mohamad Mahmoud Al Rahhal, Al Ajlan, Bazi, Al Hichri, & Rabczuk, 2018) that 12-leads ECG gives better performances in detecting Premature Ventricular Contractions (PVC) compare with the less leads ECG. The test using this ECG machine is carried out when the subject is lying still, and the 12 leads are placed on the chest, arms, and legs to sensor the electrical activity of the heart.
-
An ambulatory (Holter monitor) ECG machine: This type of ECG can take a day to two days recording ECG signals continuously. There are some arrhythmia abnormalities that rarely occur in a person, the type that may not be detected during the standard ECG testing. These kinds of arrhythmias are often tracked for 1 to 2 days using ECG holter (Takalo-Mattila et al. 2018). The electrodes may be connected to a small portable machine worn around the waist or around the hand like a wristwatch to enable monitoring of heart from home. These ECG machines are suitable for healthcare applications for remote and real time monitoring of patients.
-
Exercise ECG machine: This is a special type of ECG machine that is used for stress test. The device is usually used during an exercise and stress activity. During the exercise test, breathing and blood pressure rates can be monitored. This ECG test may be used to detect coronary artery disease, and to test safe levels of exercise following a heart attack or heart surgery.
3.2 ECG wave morphology
In the standard ECG machine with 12-leads, measurement is taken by placing the leads on the body. The leads are the channels of recording, which are lead I, lead II, lead III, aVR, aVL, aVF, V1, V2, V3, V4, V5, and V6. Among them, in particular, the lead II is most commonly used to evaluate behavior of the five waves because it shows clear signal compared to other waves (Amrani, Hammad, Jiang, Wang, & Amrani, 2018; Luo, Li, Wang, & Cuschieri, 2017; Sugimoto, Lee, & Okada, 2018). These leads are placed on the person skin usually, six on the chest. Every electrode produces the record from different angles. The resting 12-leads ECG is considered the most accurate tool to record heartbeat rhythm (Assodiky et al., 2017). However, the configuration of the ECG system used to extract signals is application-dependent (Abdeldayem & Bourlai, 2019). Some may require lying down on the bed (in the case of the resting 12-lead ECG) while some may require long time monitoring for hours to days (using Holter monitor). Methods to measure ECG have been classified as in-the-person, on-the-person and off-the-person methods. On-the-person ECG-measurement is done using electrodes attached into patient skin and this way measures the electrical activity of the heart. Secondly, device is implanted into patient’s body in-the-person measurement. Off-the-person method measures ECG without need for skin contact, for example, capacitive measurement (da Silva et al., 2015) (Muhammed & Aravinth, 2019).
The ECG signal componentsFootnote
Figure 14 shows the components of ECG waveforms recorded over an ECG machine. The ECG comprises of five (5) waves called PQRST waves. These waves give information about the electrical activities of the heart. The waves can be used for diagnosis of various heart disorders. The heartbeat is originated as an electric pulse from the sinoatrial (SA) node situated in the right atrium (singular of atria) of the heart. The SA node fires causing the atria to contract and pump blood to the lower chambers of the heart (ventricles). The P wave represents the normal atrial (upper heart chambers) depolarization, it shows how the electrical impulse (excitation) spreads across the two atriums of the heart. The Q, R and S waves that is called the QRS complex represents one single heartbeat and corresponds to the depolarization of the right and left ventricles (lower heart chambers). This occurs when the atria contract (squeeze), pumping blood into the ventricles, and then immediately relax. This is accompanied with the electrical pulse generated from the SA node which travels through the atrioventricular (AV) node that connects electrically the atria and the ventricles which activates the ventricles and cause the ventricles to contacts. The T wave represents the re-polarization (or recovery) of the ventricles. It shows that the electrical impulse has stopped spreading, and the ventricles relax once again (Antczak 2018; Banerjee et al. 2019; Swapna et al. 2018).
The structure of the human heartFootnote
3.3 Measuring of ECG waveforms and diagnoses
The reading of the normal ECG signal is done using intervals between waves. For instance, the ECG signals is presented as P-QRS-T intervals, where the P wave starts and ends before the QRS complex with a duration from 0.06 to 0.12 seconds. The PR interval has duration of 0.12 to 0.20 seconds. An extended PR interval may indicate heart blockage. The QRS complex follows PR interval with a duration from 0.06 to 0.10 seconds. ST segment prolongs from S wave to the origination of T wave. The QT interval usually appears for 0.36 to 0.44 seconds (Konan & Patel, 2018). Any extension of these intervals would indicate certain heart pathologies like arrhythmias. The American Heart Association refers to arrhythmia as any change from the normal sequence of electrical impulses. This could be slow heat beat (bradycardia) that include supraventricular tachycardia, atrial tachycardia (fibrillation and flutter), and ventricular tachycardia. Very fast heart beat (tachycardia) comprising of AV heart blocks, bundle branch blocks, and tachybrady syndrome. Irregular contraction of upper heart chamber (atrial fibrillation); abnormal heart beat (conduction disorder); early heart beat (premature contraction), and so on. Other symptoms may include fainting, dizziness, weakness, and usually pain in the chest. Sometimes, people don’t feel any symptom (Assodiky et al., 2017; Kusuma & Udayan, 2020).
However, the manual reading of ECG strips for diagnosis is time consuming and dependent on the proficiency of the Cardiologist or Physiologist. More so, it is often prone to human errors due to fatigue (Apandi et al., 2018) (A. B. A. Qayyum, Islam, & Haque, 2019). The DL have shown promising results in automatically extracting the features of the ECG raw data and analyze (Singh, Pandey, Pawar, & Janghel, 2018; Takalo-Mattila et al., 2018). This enhances the productivity of the Cardiologists by helping in making fast and accurate decisions. Other methods that are used to detect infections and conditions of the heart include chest X-ray. The Chest X-ray images have been used to detect the emerging respiratory infectious disease, also known as coronavirus 2019 (COVID-19) using DL (Ouchicha, Ammor, & Meknassi, 2020; Polat et al.; X. Zhang et al.). Although the COVID-19 is considered illness of the lungs, a study revealed that 1 in 5 patients with COVID-19 have signs of heart injury (T. Guo et al., 2020).
4 Reviews of previous surveys
It is necessary to ensure the need for a review before conducting it. The study has to starts by identifying any existing review based on the appropriate evaluation criteria on the research area (Kitchenham & Charters, 2007). In order to avoid duplication of review work, we made a general search based on the title and keywords of the current review paper and found a number of the literature reviews that survey DL applications in ECG from different perspectives (Bote-Curiel et al., 2019; Faust, Hagiwara, et al., 2018; Ganapathy et al., 2018; S. Hong et al., 2020; Rim et al., 2020; Tobore et al., 2019; Z.-J. Yao et al., 2018). A study by Ganapathy et al. (Ganapathy et al., 2018) conducted a review of DL applied in 1D biosignals such as ECG, EMG, PPG, PCG, EOG and others. This study search papers from PubMed, Scopus, and ACM databases and sampled 71 studies from 2010 to 2017 inclusive. The authors classified the DL models according to their origin, dimension, input biosignal, goal of the application, type and ground truth data; the topology of the model and the scheduling of learning. A study in (Faust, Hagiwara, et al., 2018) investigated literature works that used DL in physiological signals in healthcare applications. The survey focused on physiological signals such as EMG, EEG, ECG, and EOG. The paper extracted parameters such as specific application area, DL model, system performance and type of dataset used to develop the model. It considered 53 papers published from 2008 to 2017 inclusive. In another study, a review by Yao et al. (Z.-J. Yao et al., 2018) conducted a survey of the applications of the DL in healthcare, focusing on 7 application areas such as electronic health records (EHR), ECG, EEG, community healthcare, data from wearable devices, drug analysis and genomics analysis. The survey discussed merits and demerits of the studies identified and existing challenges before proposing future directions. The survey concluded by highlighting the robustness and interesting features that make DL suitable for clinical and healthcare data. In another survey, (Tobore et al., 2019) pointed out some biomedical domain for DL intervention in healthcare challenges. Capturing papers from PubMed and IEEE Xplore databases published between 2012 to 2017 inclusive. The study presented the applications of DL in healthcare by classifying into biological system, e-health record, medical image, and physiological signals. The survey presented research directions for enhancing health management on a physiological signal application. In a review paper conducted by (S. Hong et al., 2020), a systematic review is presented on opportunities and challenges in DL techniques on ECG data, focusing on the model architecture, the applications and dataset. The study included 191 papers from Google Scholar, PubMed and DBLP databases published from January, 2010 to February, 2020 inclusive. The authors concluded by presenting some challenges such as interpretability, scalability, efficiency as potential area for further studies. A review by Bote-Curiel et al. (Bote-Curiel et al., 2019) first presented the overview of big data and DL (which are two related fields of Data Science), in light of their applications in healthcare domain. In a two folded review, the authors reviewed applications of DL in healthcare from biomedical information with emphasis on ECG. They searched PubMed, IEEE Xplore, Google Scholar, and Science Direct electronic databases, covering papers published in 2017 and 2018. A recent review paper by (Rim et al., 2020) conducted an extensive survey on DL in physiological signal data such as EMG, ECG, EEG, and EOG. They discovered 147 papers published between 2018 and 2019 inclusive searched from PubMed database. They extracted parameters such as the input data type, task model, training architecture, and dataset sources of the DL approaches with the objective to comprehend, categorize, and compare performance as they are applied in physiological signal analysis for various medical applications.
5 Systematic literature review
To ensure knowledge advancement, literature review becomes necessary to harmonize the body of knowledge with the aim to understand, summarize, analyze, and synthesize a group of related literature (Xiao and Watson 2019). This involves evaluating, analyzing, criticizing and/or identifying missing links or research gaps. The Systematic Literature Review (SLR) utilizes the evidence-based practices of evidence-based software engineering paradigm that help in rigorous understanding of the problem domain (Babar et al. 2014). This review adopted the SLR based on the guidelines proposed by Kitchenham and Charters (Kitchenham and Charters 2007). For carrying out SLR, there are three major phases that are involved, namely, (1) Planning the review (2) Conducting the review and (3) Reporting the review.
5.1 Planning the review
Planning a SLR starts with defining the protocol or a plan that details the procedure and process in carrying out the review. It is pertinent the review protocol is meticulously checked before executing it (Brereton et al. 2007). This review protocol is pilot tested by the student and validated by the supervisors. The review protocol comprises of (a) the research questions; (b) the search strategy; (c) the inclusion and exclusion criteria (study selection criteria) and (d) the data extraction and synthesis of results.
5.1.1 Research questions (RQ)
The following questions were formulated to guide the SLR study:
RQ1: In what domains is the DL application in ECG signals presented?
RQ2: What are the DL techniques that have been applied for ECG signal analysis?
RQ3: In what application areas the proposed DL models were presented?
RQ4: What are the application tasks performed by the proposed DL models?
RQ5: What are the sources of datasets utilized in the studies to model the DL?
RQ6a: What ECG preprocessing methods and training architectures were used?
RQ6b: Which of the training architectures produced the best performance?
5.1.2 Search Strategy
The search strategy defines the strategy used to find materials for the review. These include: channels for literature search, keyword used for the search, sampling strategy, stopping rule and other restrictions (Xiao & Watson, 2019). This review searches electronic databases for published papers that applied DL in ECG signals. As suggested by Brereton et al. (Brereton et al., 2007), we selected 8 electronic databases for our literature search: IEEE Xplore digital libraryFootnote 4, ACM digital libraryFootnote 5, Science DirectFootnote 6, Springer LinksFootnote 7, DBLPFootnote 8, PubMedFootnote 9, and two interdisciplinary research databases, ScopusFootnote 10 and Web of Science (WoS)Footnote 11. Selective and representative sampling was adopted in this review, with only peer-reviewed studies based on application of DL in ECG. The papers included in the literature were those written in English language only. The search covers from January 1st, 2010 to April, 30th 2020 (current date of conducting the research) inclusive. The following keywords were used; “deep learning”, “deep neural network”, “deep neural networks” “convolutional neural network” and “Electrocardiogram”, “ECG”, “EKG” (S. Hong, Zhou, Shang, Xiao, & Sun), and “deep learning electrocardiogram ECG” (Rim et al., 2020). Boolean operators were used to concatenate the keywords. The “OR” operator is often used to join synonymous keywords while “AND” operator is used to combine the main terms in the search string (Brereton et al., 2007). We used Boolean operators because most of the databases support their use (Xiao & Watson, 2019). These keywords were integrated to form the search string as shown below.
(“DL” OR “deep neural network” OR “deep neural networks”) AND (“Electrocardiogram” OR “ECG” OR “EKG”) AND DL electrocardiogram ECG
The initial search string was tested on two databases, Science Direct and IEEE Xplore by way of validation, the string fetched relevant known and unknown primary studies. In some cases, the search string had to be modified to suit the databases.
5.1.3 Study Selection Criteria
In view of the research questions and SLR objectives, the following inclusion and exclusion criteria were defined and observed in the published papers that were retrieved.
-
A.
Inclusion criteria.
Primary studies that contain the following will be included:
-
Study that presents evidence of the use of a DL architecture to model ECG AND.
-
Study based on empirical evidence AND.
-
Study that is reported in peer-reviewed workshop or conference or journal AND.
-
Study written in English Language AND.
-
Study between January 1st 2010 and 30th April 2020 inclusive.
-
B.
Exclusion criteria.
A primary study that contains the following will be excluded:
-
Study on deep model architecture without evidence of application in ECG OR.
-
Study not accessible electronically OR.
-
Study that is not complete (content and incomplete results) OR.
-
Encyclopedia, posters, books, book chapters, keynotes, and editorials OR.
-
Study that is a duplicate (if two versions of a paper are found, the less complete version is excluded).
-
C.
Quality Assessment (QA).
The QA was applied on the papers that passed the inclusion criteria. This is the final stage for preparing the selected papers for data extraction and analysis stage (Xiao & Watson, 2019). The full texts of the papers were read and checked considering the QA checklist presented in Table 1.
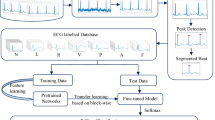
The QA was formulated based on the research questions and as suggested by Kitchenham and Charters (Kitchenham & Charters, 2007). Each criterion is computed using the following scale: Yes (Y) = 1 point, No (N) = 0 point; and Partial (P) = 0.5 point. The scores were summed to determine whether the paper is fit for inclusion for data extraction and analysis stage. The score (x) for each study lies between 0 < = x => 10 points. The cutoff point is computed by taking the quartile (10/4 = 2.5), this implies that only studies that score x > 2.5 would be included for our final list of primary studies. At the end, 150 papers passed the threshold value and were included for data extraction and analysis (see Fig. 16).
5.1.4 Strategies for data extraction and synthesis
To aid in the data extraction and synthesis, a form was designed to include information to be extracted such as the publication details, titles, authors and year. The fields were provided for the application domain, ECG preprocessing method, application task, DL model, DL model performance, training architecture, datasets and number of subjects/data used. These data correspond to the research questions and consequently helped in answering of the research questions as well.
5.2 Conducting the review
This section reports the process of how the search for papers for this SLR was conducted. The search strings were applied on the databases to obtain potential primary studies for inclusion in the SLR. Figure 16 clearly illustrates the stages followed to obtain our final papers.
The process in the selection of papers, n, on all the identified search results (n = 2886) in light of the inclusion and exclusion criteria involves two phases: reading through the title, keywords and abstract of the paper. To avoid researcher bias, two researchers were involved. The selection process was performed by the first author and checked by the supervisor. At the end of this screening, (n = 873) papers were included for the next step. The papers were then checked for duplicates for each of the screened papers. This resulted to (n = 492) papers screened for the next phase. In the second phase, full text reading was performed by the first author for eligibility, who read through all the unduplicated papers. However, discrepancies in the assessment results were discussed between the first author and the supervisor and resolved accordingly. Finally, (n = 150) papers were included for data extraction, including 5 papers added using manual search (see Fig. 16).
Figure 17 (a) shows a steady rise of number of papers with the year 2019 having the highest number with 55 (36.7%), followed by 2018 (47, 31.3%), then 2017 (23, 15.3%). In 2020, 17 (11.2%) papers were published (the low number of papers in 2020 can be explained from the fact that the search was conducted in April 2020). There were 6 (4%) papers retrieved from 2016, 1 (0.8%) from 2015 and from 2014, respectively. Based on the included papers, no paper reported application of DL in ECG signals from 2010 to 2013. This is likely because DL was not yet popularly used as illustrated on the Google trends as shown in Fig. 1.
Figure 17 (b) shows the type of venue the papers were published categorized as conference papers, journals and workshops or symposiums. 58 (38.7%) papers were published conference papers, 89 (59.3%) were journals while 3 (2%) were published workshops papers.
5.3 Reporting findings
This section reports the results of the SLR. This involves the presentation of findings in the extracted papers and answering of the research questions formulated in section 5.1.1.
5.3.1 Application of DL in ECG signal
In this section, we categorized the application of DL in ECG data into domains of application.
RQ1: In what domains is the DL application in ECG signals presented?
Research question 1 seeks to present the domain of the DL application in ECG data as reported in the included papers. Based on the study systematic review, the applications of the DL in ECG signals have been categorized into three domains of applications: Medical/healthcare domain; Biometric/security domain and Driving domain.
5.3.1.1 Medical/Healthcare domain
The DL has become an active area, vital in many medical and healthcare applications to enhance the quality of diagnosis. The major role of healthcare personnel is to diagnose a disease and find the best means for suitable treatment. This comes with challenges and responsibilities for the healthcare practitioner for years. In addition, a medical doctor is a professional who makes intelligent decisions based on symptoms and test results. However, to achieve good decisions some sort of knowledge is necessary (Faust, Hagiwara, et al., 2018). The ECG plays an essential role in diagnosing and screening CVDs that helps medical Doctors and Cardiologist to make more informed decisions. DL is being utilized in physiological signals to discover hidden relationships that help medical practitioners and healthcare providers to make fast and informed decisions and to predict numerous clinical events (Yang, Islam, & Li, 2018).
The DL have shown the potential to improve the accuracies of CVDs diagnosis such as arrhythmia detection using ECG signals (Ding et al., 2019). A discussion of the applications of DL in cardiology is presented in (Bizopoulos & Koutsouris, 2018). A review conducted by Kusuma and Udayan (Kusuma & Udayan, 2020) reveals that DL is the attractive algorithm for CVDs diagnosis. This section summarizes the papers that are found to apply DL in the medical and healthcare applications for ECG modeling, with emphasis on the application models, application task, sources of data, performances achieved by the proposed DL models and limitations of the studies.
-
I.
Deep Neural Networks.
This section discusses DNNs based models proposed in the literature for automatic classification of the ECG signals.
Arrhythmias detection using Genetic Algorithm (GA) and Particle Swarm Optimization (PSO) as features extractors from ECG signals and performed classification using DL was proposed in (Assodiky et al., 2017). Using 452 data samples from UCI-ML Repository, model performance was indicated that PSO have the highest accuracy of 76.51%. It was also shown that PSO reduces features of 261 to 23 attributes whereas GA reduced to 31 attributes. Although, the number of normal ECG signals classes were higher than the other classes. A study conducted in (Jeon, Chae, Han, & Lee, 2019) and (Jun, Park, Minh, Kim, & Kim, 2016) proposed a DNN models for arrhythmia classification and premature ventricular contraction (PVC) beat, respectively. The result indicated accuracy of 98.07% and 99.41% respectively using MIT-BIH Arrhythmia Database (MITDB). However, there was limited dataset for training and testing the models. In a study by Sannino and De Pietro (Sannino & De Pietro, 2018), the authors proposed a DNN model that utilized temporal feature extraction and classified ECG signals. A good accuracy of 99.68%, sensitivity of 99.48% and specificity of 99.83% were obtained based on evaluation on the MITDB. Though, there was too much experts’ involvement in the ECG beats annotation process. Arrhythmia classification was performed based on DNN model proposed by Xu et al. (shensheng Xu, Mak, & Cheung, 2017). The study investigated the effect of preprocessing methods. A method called Fisher discriminant ratio (FDR) produced the best performance with accuracy of 82.96% evaluated using UCI cardiac arrhythmia dataset. However, there was limited data and unbalanced data problem. In a study conducted by Xu (S. S. Xu, Mak, & Cheung, 2018) an end-to-end DNN model to classify ECG beats was proposed. The study evaluated their model based on expert intervention and without expert intervention. The results show that a better results without expert intervention on S and V class of ECG beats evaluated on the MITDB was achieved. However, there was limited data and imbalance data problem.
Note: Acc = Accuracy, Spe = Specification, Sen = Sensitivity, Pre = Precision, Ppr = Positive productivity
A study by Cai et al. (Cai et al., 2020) proposed a model based on deep densely neural networks (DDNN) to detect Atrial Fibrillation (AF). In the study, only AF class was considered due to insufficient data in the other classes, the performance in terms of accuracy, specificity and sensitivity range above 99% using three dataset sources of 12-leads ECG including the China Physiological Signal Challenge 2018, the Chinese PLA General Hospital and wearable ECG devices from CardioCloud Medical Technology (Beijing) Co. Ltd, with 11,994 unique subjects. A Prenatal Detection of Congenital Heart Disease (CFD) was proposed by Vullings (Vullings, 2019). The study used a DNN model to detect CFD with 76% accuracy using private dataset containing fetal ECG measurements from 266 healthy and 120 CHD groups. The performance may be improved with more population or dataset. Table 2 presents the summary of the study on DNN for ECG signal processing.
-
II.
Convolutional Neural Networks.
In this section, we discussed CNN-based models proposed for ECG classification purposes. CNNs have shown promising performances especially in the area of image recognition (Kulik & Shtanko, 2020) and computer vision(Khan et al., 2018; Voulodimos et al., 2018). And it is considered the most used DL especially for medical time series data and imaging (Pourbabaee et al., 2018; Shi, Wang, Qin, Zhao, & Liu, 2020). Table 3 summarizes the applications of CNN-based models for heartbeats ECG signals analysis.
A study by (Acharya, Fujita, Lih, Hagiwara, et al., 2017) proposed a CNN model to detect different arrhythmias. The study proposed two CNN architectures (Net A and Net B) with 500 input and 1250 input samples respectively. The performance of the two proposed CNNs in terms of accuracy, specificity and sensitivity is greater than 90% respectively with exception of Net B that achieved 81.44% accuracy using the Creighton University Ventricular Tachyarrhythmia database (CUDB), MIT-BIH atrial fibrillation database (AFDB) and MIT-BIH arrhythmia database (MITDB). The DL architectures demonstrated good performances with 2 and 5 s ECG segments respectively. However, there was limited data for training and too much training time was observed. Data augmentation and bagging algorithm could solve limited data problem. The classification of shockable and non-shockable life-threatening ventricular arrhythmias using 2 s ECG segments was proposed by (Acharya et al. 2018) using a CNN architecture. For immediate treatment of shockable ventricular arrhythmias, cardiopulmonary resuscitation (CPR) and defibrillation have been used and highly recommended. However, To improve the efficiency of automated external defibrillator (AED) for defibrillation, accurate diagnosis of these shockable and non-shockable ventricular arrhythmias is necessary. The CNN model achieved 93.18% accuracy evaluated using MITDB, VFDB and CUDB. However, limited data and too much training time problems affected the performance. In a study by (Acharya, Oh, et al., 2017) the study proposed a CNN model to classify heartbeats and it obtained accuracy of 94.03% (Set B) using the MITDB. A study conducted by (Al-Huseiny et al. 2020) proposed a 2D-CNN model for arrhythmias recognition. They first converted the ECG signals into 2D ECG images which served as input to the training model. The model achieved accuracy of 96.69% on the MITDB.
There are CNN-based models that have been trained using large scale data, these models can be reused with no or minimal tuning using either the same (source data) or different type of data (target data). These models are called pre-trained models (also known as transfer learning models) because they have been trained using a large scale data (Ghaffari and Madani 2019). These models are used largely when there is insufficient data and hardware to train the target data (Byeon et al. 2020). A study in (Alquran et al. 2019) utilized two pre-trained CNN models namely GoogleNet and AlexNet to classify ECG signals. Higher Order Spectral estimation was first applied on the ECG signals to extract features, that were fed into the pre-trained models for classification. The accuracy of 97.8% was achieved using third cumulants + GoogleNet using the MITDB. Another pre-trained CNN called VGG-Net was proposed by (Mohamad M Al Rahhal, Bazi, Al Zuair, Othman, & BenJdira, 2018) for end-to-end classification of ECG signals. Good performances were recorded in terms of ventricular ectopic beat (VEB) and supraventricular ectopic beat (SVEB) evaluated using the MITDB. (Amrani et al. 2018) proposed a very deep CNN model as feature extractor using a fusion technique called multi-canonical correlation analysis (MCCA), and the extracted features were classified using Q-Gaussian multi-class SVM (QG-MSVM) (Amrani et al. 2018). The effect of the fusion technique was observed to outperform the compared methods with 97.37% accuracy for arrhythmias detection analysis. The fusion technique may introduce additional training time. A study in (Brito et al. 2019) proposed pre-trained based CNN model, ResNet. The model was optimized using stochastic gradient descent (SGD) and adaptive moment estimation (Adam) with 0.1 learning rate respectively. The experiments show that the SGD attained higher accuracy of 96% compared with 83% obtained using Adam. However, the imbalance of data can be investigated further to improve performance. The effect of adding a convolutional block on the baseline CNN (network A) consisting of three convolutional layers (network B) called a multi-scale fusion CNN was propose in (Dang, Sun, Zhang, Zhou, et al., 2019). The proposed model (Network B) achieved average performance of 95.48%, 96.53%, and 87.74% for accuracy, sensitivity and specificity, respectively, evaluated using the MITDB. A study by (Diker and Engin 2019) proposed a model combining CNN and Extreme Learning Machine (ELM) for classification of ECG signals. Although the performance accuracy was below 90% (88.33%) using Physikalisch-Technische Bundesanstalt Diagnostic database (PTBDB), yet was found to be better compared with the traditional models such as K-NN, Decision Tress and SVM. A study conducted by (Dokur and Ölmez 2020) proposed a heartbeat classification model based on CNN, excluding the fully connected neural network (FCNN) part of the basic CNN model. However, a Walsh functions was applied to maintain performances during training. Also, the drawback of converting one-dimensional (1D) ECG signals to two-dimensional (2D) images was investigated. The average accuracy of 99.45% and 98.7% for 1D ECG signals and 2D ECG images, respectively, were achieved when evaluated using the MITDB. In a study by (Fujita and Cimr 2019) proposed recognition of ECG rhythms application based on CNN. The model first used continuous wavelet transformation (CWT) to extract features, which was fed into the CNN architecture for automatic classification of the ECG rhythms. An accuracy of 97.78% was achieved.
However, knowledge of features is required before the training model. A multi-channel CNN model was proposed by the Hao et al. (Hao et al. 2019) for ECG beats classification. The authors used beat-to-beat and single-beat information. At first, it converted ECG signals using short-time Fourier transform (STFT) and wavelet transform to obtain spectro-temporal images that were fed into the model for training. The model obtained a comparable detection performance based on sensitivity and positive predictive value (PPV). Also, a study by Huang et al. (Huang et al. 2019) proposed 2D-CNN model which was used to train ECG images transformed using STFT. The model was simulated on the MITDB and obtained accuracy of 99.00% for ECG classification task over the ID-CNN.
Cardiac arrhythmia detection study was conducted by Isin and Ozdalili (Isin and Ozdalili 2017) using pre-trained CNN model (AlexNet) as feature extractor. The model was trained using back-propagation algorithm (BPNNs). A recognition rate of 98.51% was achieved on the MITDB. A 2D-CNN model was proposed by Izci et al. (Izci et al. 2019) for arrhythmia detection task. A performance accuracy of 97.42% was achieved on the MITDB. A Deep residual CNN model was proposed by Kachuee et al. (Kachuee et al. 2018) for Arrhythmia classification. The model was used for myocardia infarction (MI) prediction. The performance achieved was 93.4% accuracy for arrhythmia classification, and 95.9% accuracy for MI prediction on the PTBDB. A CNN model was proposed by Kaouter et al. (Kaouter et al. 2019) for ECG classification task. The model was compared with Ensemble of Fine-Tuned CNN, VGG Net-16 and Res Net-50 and CNN full training, Google Net-144 layers was found to produce the best performance accuracy with 93.75% using ECG signals evaluated on the MITDB, MIT-BIH normal sinus rhythm database (NSRDB) and Beth Israel Deaconess Medical Center (BIDMC) congestive heart failure database. However, the diagnostic value of the proposed model can be improved by including other parameters of patients other than the ECG signals. Real time ECG classification task was proposed by Kiranyaz (Kiranyaz et al. 2015). The model was evaluated using the MTIDB and obtained a good performance on VEB and SVEB. However, there was limitation on the characterization of crucial beat like S beat. More so, the proposed model does not support learning.
Also, (Li et al. 2017) conducted a study for ECG classification using 1D-CNN. The model achieved accuracy of 97.5% evaluated using the MITDB. More ECG leads can be investigated for better detection. A pre-trained model (ResNet-31) was proposed by Li et al. (Li et al. 2020) for ECG classification. The model was tested on 2-lead and single ECG data. The result on 2-lead data achieved the best performance with 99.38% accuracy over the single lead data with 99.06% accuracy. However, it takes long time to converge during training on the 2-leads ECG dataset than the single ECG dataset. In a study by Pandey and Janghel (Pandey and Janghel 2019), it proposed a CNN-based model for arrhythmia detection problem. Because of the imbalance in the MITDB, Synthetic minority oversampling technique (SMOTE) technique was used to solve the imbalance problem. The model achieved a good performance with 98.30% accuracy, 86.06% precision, 95.51% recall and 89.87% F1-score. A study by Pyakillya et al. (Pyakillya et al. 2017) proposed a 1D-CNN model for ECG signal classification task. The study was evaluated on the PhysioNet/Computing in Cardiology Challenge 2017 (PhysioNet/CinC 2017) dataset and obtained an accuracy of 86%. The CNN model, however, imbalance dataset is used. The authors proposed to use GANs to solve the imbalance dataset problem. A CNN model was proposed by Rajkumar et al. (Rajkumar et al. 2019) for Arrhythmia classification task. The CNN model obtained accuracy of 93.6% on the MITDB. A recent study conducted by Ribeiro et al. (Ribeiro et al. 2020) proposed a residual framework (ResNet), for effective diagnosis of ECG using 2, 322, 513 ECG records of private dataset collected from 1,676,384 different patients. Using the short-duration and standard 12-lead ECG, the model achieved F1-score greater than 80% and specificity greater than 99%. The ECG classification task using deep CNN and two-stage deep CNN was proposed by Shaker et al. (Shaker et al. 2020). The authors also demonstrated the effectiveness of heartbeat augmentation using GANs which yielded better performances compared with the unbalanced data. The models achieved overall accuracy > 98.0%, precision > 90.0%, specificity > 97.4% and sensitivity > 97.7%. However, post-processing method such as smoothing filter can be applied to enhance heartbeat quality. Also, outlier removal can enhance precision results. A CNN model was proposed by Xu and Liu (Xu and Liu 2020) and it achieved a good performance with average accuracy of 99.43% on both SVEB and VEB beats evaluated on the MITDB. A Multi-Scale CNN (MCNN) was proposed by Yao and Chen (Z. Yao & Chen, 2018). Although the study used single lead ECG as input without considering rhythm information, the model achieved performance better than the methods that use hand-crafted features with 88.66% overall accuracy on SVEB and VEB. Landmark information can be utilized to improve performance of the model.
A study conducted by Yıldırım et al. (Yıldırım et al. 2018) used 1D-CNN for arrhythmia detection. The model used the MITDB to classify the ECG and achieved 91.33% accuracy, 83.91% sensitivity and 99.41% specificity. A CNN model proposed by Yu et al. (Zhang et al. 2018) for ECG classification achieved accuracy of 98.92%, sensitivity of 98.37% and specificity of 99.19% on the MITDB. The CNN-based architecture was utilized for feature extraction in a study proposed by Zhou and Tan (Zhou and Tan 2020). The authors used extreme learning machine (ELM) for the classification of the ECG and achieved 98.77% accuracy on the MITDB. A study conducted by Li et al. (Li et al. 2018) proposed a novel approach by fusing the morphology and rhythm of heartbeats to feed into a CNN model for ECG classification. The authors used one-hot encoding technique to convert the 1D ECG signals to 2D images to improve the convergence speed and improve the accuracy. The model was tested on the MITDB and achieved a performance of more than 90% accuracy on both SVEB and VEB. However, the specificity and accuracy of V beat was lower than the compared methods because of the use of raw ECG without representation extraction. A study by Takalo-Mattila et al. (Takalo-Mattila et al. 2018) proposed a 1D-CNN model to classify ECG which was tested on the MITDB and achieved a competitive performance on the Normal, SVEB and VEB classes.
Automatic heartbeats disease classifications such as myocardial infarction disease (MID), Anterior Myocardial Infarction (AMI), congestive heart failure (CHF), Atrial Fibrillation (AF) etc. have been classified using CNNs based models. The MID is also known as heart attack. It occurs due to the damage of heart muscle, causing the decrease of blood flow (Yufei Chen, Chen, He, Yang, & Cao, 2018). A study by Acharya et al. (Acharya, Fujita, Oh, et al., 2017) proposed a CNN model for automatic detection of MI. The study demonstrated the effect of noise by removing baseline wander in the ECG signals in the first dataset and kept the noise on the other dataset. The result shows that the performance recorded were 93.53%, 93.71% and 92.83% for accuracy, sensitivity and specificity, respectively (using dataset with noise). The de-noised dataset obtained better performance with accuracy of 95.22%, sensitivity of 95.49% and specificity of 94.19%, all evaluated on the PTBDB. Another study by Acharya et al. (Acharya et al. 2019) proposed a CNN model for the diagnosis of CHF. The study evaluated the model based on the combination of databases into set A, B, C and D. Set A (NSRDB, BIDMC), Set B (Fantasia, BIDMC), Set C (NSRDB, BIDMC) and Set D (Fantasia, BIDMC). The CNN model achieved accuracy, sensitivity and specificity greater than 90% in each of the scenarios. A study by Ahmed et al. (Ahmed et al. 2019) proposed a CNN model to predict MID, however, the authors optimized the CNN model using Ant Colony Optimization (CNN-ACO). An accuracy of 95.78% was recorded on the UCI-ML Repository. The CNN-ACO model, however, consumed a lot of memory compared with the basic CNN model, likely because of the additional ACO. The automatic detection of MI was proposed by Alghamdi et al. (Alghamdi et al. 2019) using a pre-trained deep CNN (VGG-Net). Two architectures of VGG-Net (Fine-tuning and VGG-Net as fixed feature extractor) were proposed, namely, VGG-MI1 and VGG-MI2. The VGG-MI1 is based on the VGG-Net model with little fine-tuning. And VGG-MI2 was used as the feature extractor and QG-MSVM as the classifier. It improve the accuracy by 2%. VGG-MI2 obtained the best accuracy of 99.22% on the PTBDB. A study proposed by Baloglu et al. (Baloglu et al. 2019) used deep CNN model to classify MI. Result indicated that the accuracy of 99.78% was achieved on PTBDB. However, the model did not detect the locations of the MI.
A Multi-Channel Lightweight CNN (MCL-CNN) was proposed by Chen et al. (Chen et al. 2018). The model was tested on PTBDB and achieved accuracy of 96.18% for detecting Anterior MI (AMI). A study by Erdenebayar et al. (Erdenebayar et al. 2019) proposed a 1D-CNN model for automatic prediction of AF. Result shows 98.7% accuracy, 98.6% sensitivity and 98.7% specificity were recorded on the AFDB, Paroxysmal Atrial Fibrillation Prediction Challenge Database (PAF-DB) and NSRDB. However, the model could not detect the starting point of AF and only potential AF subjects were analyzed. The AF identification study was put forward by Ghaffari and Madani (Ghaffari and Madani 2019) using pre-trained CNN models (AlexNet, VGG-16, ResNet-152 and Alexnet-scratch), 1D-CNN as feature extractors, MLP and SVM as classifiers. The AlexNet + MLP model achieved the best performances with accuracy of 87.6% (87.9%), sensitivity of 81.1% (85.7%), and specificity of 94.3% (92.7%) (the noisy classes in parenthesis). The model was evaluated on the PhysioNet/CinC Challenge 2017. Another study by Hsieh et al. (Hsieh et al. 2020) proposed 1D-CNN for AF diagnosis. The model was tested on PhysioNet/CinC Challenge 2017 and achieved 90.7% accuracy for AF detection. Classification of ECG signals into AF and normal sinus rhythm was proposed by Huang and Wu (M.-L. Huang and Wu 2020). The study proposed a 2D-CNN trained on the AFDB and NSRDB Dataset. The study was evaluated using filtered ECG signals (experiment 1) and non-filtered ECG signals (experiment 2). Experiment 1 achieved the best performance with accuracy of 99.23%, sensitivity of 99.71% and specificity of 98.66%. A study conducted by Kamaleswaran et al. (Kamaleswaran et al. 2018) proposed a deep CNN model for AF detection tasks and achieved accuracy of 85.99% and F1-score of 0.83 on PhysioNet/CinC Challenge 2017. A study by Li et al. (Li et al. 2018) proposed CNN model for feature extraction and used MLP for classification. The model achieved accuracy of 93.14%, sensitivity of 83.5% and specificity of 95.99%. However, because at the time of the study there was no public available dataset on postoperative, detailed information for patients with AF, they acquired their dataset using 128-Leads BSPM System from West China Hospital on 14 AF patients. Also, (Li et al. 2019) combined CNN and SVM for AF classification. The model classified the ECG signals using SVM fed with features extracted using CNN architecture. Though, the model achieved 96% accuracy, 88% sensitivity and 96% specificity, but the data used was too small.
MI detection and localization study was proposed by Liu et al. (Liu et al. 2018). The study proposed a novel Multiple-Feature-Branch CNN (MFB-CNN) model in which each branch corresponds to a certain lead. The model was tested on PTBDB based on class-based scheme and patient-specific scheme. Class-based achieved accuracy of 99.95% on detection and 99.81% on localization while patient-specific achieved accuracy of 98.79% on detection and 94.82% on localization. A study for detection of fetal ECG was proposed by Lo and Tsai (Lo and Tsai 2018). With the help of non-evasive abdominal ECG recording, fetal ECG (fECG) is made easy to monitor. A STFT was first applied on the fECG signals to obtain the time-frequency representation that was fed into the 2D-CNN for automatic detection. Abdominal ECG recording from PhysioNet database was used to test the model. It found that it achieved 92.65% accuracy. A combination of CNN and conventional classifiers (KNN, MLP and SVM) were proposed by Pourbabaee et al. (Pourbabaee et al. 2018) for screening paroxysmal atrial fibrillation (PAF) task. Using the PAF prediction challenge database, the best model combination (CNN + KNN) achieved the best performances. The classification of AF task was proposed by Qayyum et al. (Qayyum et al. 2018) using pre-trained models namely, AlexNet, VGG16, VGG19, GoogLeNet and ResNet-101. The models were used as feature extractors with combination of ensemble classifier and SVM as classifiers. However, the ResNet-101 model was only fine-tuned and used as end-to-end classifier. The ResNet-101 achieved the best performances with accuracy of 97.89%, sensitivity of 97.12% and specificity of 96.99% when tested on PhysioNet/CinC Challenge 2017 dataset. DenseNet, a pre-trained CNN-based model was proposed by Rubin et al. (Rubin et al. 2018) for AF detection task. The model was tested on the PhysioNet/CinC challenge 2017 dataset and obtained F1 score of 0.82. A study by Xia et al. (Xia et al. 2018) proposed a CNN model in an end-to-end classification of AF. The model received an input from the transformed ECG segments by STFT and stationary wavelet transform (SWT) respectively. The models were evaluated on the AFDB and achieved competitive performance with sensitivity of 98.34%, specificity of 98.24% and accuracy of 98.29% (STFT-CNN). On the other hand, SWT-CNN model achieved sensitivity of 98.79%, specificity of 97.87% and accuracy of 98.63%. AF detection was studied by Xiong et al. (Xiong et al. 2017) using CNN model. The result of the CNN is compared with the RNN and spectrogram learning. The proposed CNN model outperformed the compared algorithms with 82% over all accuracies. However, imbalance ECG data and varying lengths affected the performance of the model.
There are health challenges associated with sleep disorder which have been investigated. Among which include obstructive sleep apnoea (OSA) or sleep apnoea syndrome (SAS). Polysomnography (PSG) is the standard method for sleep apnoea diagnosis. However, because of the cost and time consumption involved in the PSG, recently, studies move toward effective classification of sleep apnoea based on ECG signals. Sleep apnoea classification based on CNN model was proposed by Dey et al. (Dey et al. 2018). Using the apnoea-ECG dataset from Physionet, the model achieved an accuracy of 98.91%. However, mostly expert observation is done in offline process. A study proposed by Urtnasan et al. (Urtnasan et al. 2018) applied CNN model for the classification of multiclass obstructive sleep apnea/hypopnea. The ECG signals were measured by a single-lead transducer at lead II during the nocturnal PSG. The test results obtained accuracy of 90.8%, sensitivity of 87.0% and specificity of 87.0% with 87.0 precision, recall and F1-score. In another study, detection of OSA was proposed by Urtnasan et al.(Urtnasan et al. 2018) using a CNN model. The test results produced accuracy, sensitivity, specificity of 96.0% as well as 0.99 precision, recall and F1-score. However, these studies did not provide the information on the cause-based classification of sleep apnea for central apnea and mixed apnea. Also, ECG recordings were not completely independent from other events, such as snoring, movement, and flow. More so, only one specialist was involved in the annotation and segmentation without cross-checking of events. A study applied CNN model for classification of Coronary Artery Disease (CAD) (Acharya, Fujita, Lih, Adam, et al., 2017). The authors divided the dataset into two, 2 s segments of ECG (Net A) and 5 s ECG segments (Net B) which they used to classify two classes of ECG signals (normal and CAD). The performance obtained for both the two sets of the proposed CNNs structures were 95% (Net A) and 95.1% (Net B), evaluated on the Fantasia database and St.-Petersburg Institute of Cardiology Technics.
The classification of stress into different levels has been a challenging task. A study by Giannakakis et al. (Giannakakis et al. 2019) proposed stress recognition using 1D Deep Wide CNN (DWNet1D) model. They utilized 4 different stressors in a bit to presents different stressors scenarios. The dataset was collected during the data acquisition campaign (SRD’15) from 24 subjects. The model achieved accuracy of 99.1%. However, limited data was used in the study and the problem of inter-subject variability could affect the performance of the model. The summary of the studies that apply CNN for ECG is presented in Table 3.
-
III.
Recurrent Neural Networks.
Applications of RNNs-based models in ECG have been proposed in the literature. A study presented in (Banerjee et al. 2019) proposed RNNs-based model which combines two LSTM networks and the output was merged with hand-crafted features of ECG signals to classify the AF. The proposed model obtained sensitivity of 0.93, specificity of 0.98 and F1-score 0.89. This model, used hand-crafted features. MI classification was proposed by Darmawahyuni and Nurmaini (Darmawahyuni and Nurmaini 2019) and used LSTM model to perform binary classification of MI. The model achieved precision of 0.91, sensitivity of 0.91, F1 score of 0.90 and BAcc of 0.83 using PTBDB. In another study, Darmawahyuni et al. (Darmawahyuni et al. 2019) proposed RNN, LSTM and GRN models for MI classification. Among the proposed RNNs models, LSTM produced the best results with sensitivity of 98.49%, specificity of 97.97%, precision of 95.67%, F1-score of 96.32%, BACC of 97.56%, and MCC of 95.32% on PTBDB.
Detection of AF using bi-LSTM was proposed by Faust et al. (Faust, Shenfield, et al., 2018). The model yielded accuracies, specificities and sensitivities greater than 98% for both cross and blind validation. A study presented by Sujadevi et al. (Sujadevi et al. 2017) proposed RNN, LSTM, and GRU models for AF detection. Without any preprocessing method applied, the models achieved accuracy of 0.950, 1.000 and 1.000, respectively using AFDB and NSRDB from MIT-BIH Physionet. A LSTM model was proposed by Chang et al. (Chang et al. 2020) for the detection and classification of the cardiac arrhythmias. The model was found competitive compared with Cardiologists, emergency physicians and internal-medicine doctors. The model achieved 90% accuracy using ECG signals collected from the China Medical University Hospital (CMUH) recorded by a GE Marquette MAC 5500. However, the ST-T change which is important for diagnosing acute myocardial infarction was not detected. A study presented by Deshmane and Madhe (Deshmane and Madhe 2018) proposed LSTM model for detection. Results show that precision of 0.91, sensitivity of 0.91 and F1 score of 0.90 were recorded. The performance of this model can be improved upon by providing a more robust preprocessing method. Arrhythmia recognition was proposed by Pandey and Janghel (Pandey and Janghel 2020). Morphological, statistical, R-R interval and wavelet features were fed into LSTM for the classification. The model achieved precision of 96.73%, accuracy of 99.37%, specificity of 99.14%, F-score of 95.77%, and sensitivity of 94.89% on the MITDB. However, the model used hand-crafted features. A study presented by Sharma et al. (Sharma et al. 2020) proposed a LSTM model for arrhythmia classification. Fourier-Bessel expansion was used to derive the intelligent series from the RR-intervals and was fed into the LSTM model for classification. The model achieved an accuracy of 90.07% and 89.04% on MITDB and a private dataset, respectively.
A study conducted by Saadatnejad et al. (Saadatnejad et al. 2019) proposed a LSTM model for ECG classification. The model was fed with RR features and wavelet features for training. The LSTM model was evaluated on the MITDB and achieved accuracy of 99.2% and 98.3% for VEB and SVEB, respectively. A study proposed by Singh et al. (Singh et al. 2018), presented a study on RNNs based models (RNN, GRU and LSTM). An accuracy of 85.4% (RNN), 82.5% (GRU) and 88.1% (LSTM) was achieved when tested on MITDB. However, hand-crafted features were use and it was not clear if the features will work on a new disease. A study presented by Yildirim (Yildirim 2018) proposed a deep bidirectional LSTM-based model for ECG classification. This model proposed a wavelet-based layer to improve the classification performance using wavelet sequences (WS). The proposed DBLSTM-WS obtained the best performance when WS layer is 3, with 99.39% on the MITDB. However, due to the hardware limitation not all dataset from the MITDB was used. A study based on LSTM model was conducted for OSA detection (Cheng et al. 2017). The model was tested on the Apnea-ECG database, having 70 ECG record and obtained detection accuracy of 97.80%. The summary of the studies that applied RNN and its variants is presented in Table 4.
-
IV.
Restricted Boltzmann Machines.
In this section, studies based on RBMs for ECG modeling are presented. From our review, few studies were found to be based on the RBMs for ECG modeling. A study for arrhythmia detection was proposed by Altan et al. (Altan et al. 2018) using a multi-stage DBN model; a greedy layer-wise unsupervised and supervised training RBM-DL. Techniques such as higher order statistics, morphology, Wavelet packet decomposition and Discrete Fourier transform were used for featured extraction. The model achieved performance of 94.15% accuracy, 92.64% sensitivity and 93.38% selectivity on MITDB. However, the comparison was difficult due to different number of heartbeats. RBM-DBN model was proposed by Mathews et al. (Mathews et al. 2018) for ECG classification. The proposed model was evaluated on the MITDB and achieved accuracy of 93.78% (SVEB) and 96.94% (VEB) on the sampling rate of 360 Hz. And using the sampling rate of 114 Hz, accuracy of 93.63% (VEB), and 95.57% (SVEB) was achieved. A study by Wu et al., (Wu et al. 2016) proposed a RBN model for ECG classification of arrhythmia. Two types of RBM; Bernoulli–Bernoulli and Gaussian–Bernoulli were stacked to form DBN. The model achieved accuracy of 99.3% on S class and 97.9% on V class, respectively on MITDB. Another RBN model was proposed by Mostafa et al. (Mostafa et al. 2017) for sleep apnea detection. The model achieved accuracy of 85.26% on the UCD database and 97.64% on the apnea-ECG database. However, the model was tested with imbalanced dataset. A study in (Huanhuan and Yue 2014) proposed two DL based models based on DBNs; DBN + NNs and DBN + SVM. The best performance of 98.49% accuracy was achieved by DBN + SVM with Gaussian kernel. However, PCV and APB recognition performance of proposed scheme was poor. The summary of the studies on RBM for ECG is presented in Table 5.
-
V.
Autoencoders.
This section presents the application of DL in ECG signals based on AEs models found in this literature review study. An AE-based deep network was proposed by Debnath et al. (Debnath, Biswas, Ashik, & Dash, 2018) for classification of cardiac arrhythmia groups. The proposed model achieved 92.1% accuracy on the MITDB. A study conducted by Al Rahhal et al. (Mohamad Mahmoud Al Rahhal et al., 2016) proposed a stacked denoising autoencoders (SDAE) model to extract features from the ECG signals for active ECG classification. The model was evaluated on the benchmark arrhythmia database (MITDB) as well as INCART and SVDB and achieved very good performances on VEB and SVEB. However, experts were allowed to label the most relevant and uncertain ECG beats in the test record during the model training. A study presented by Luo et al. (Luo et al., 2017) proposed SDAE model for patient-specific ECG classification. The authors first converted the raw ECG signals into time-frequency image using modified frequency slice wavelet transform (MFSWT) which was fed into the SDAE for feature extraction. The model used the encoder layer of SDAE and a softmax layer to form a DNN model for the classification. The model produced classification accuracy of 97.5% when evaluated on the MITDB.
Classification of AF based on stacked AE model was proposed by Farhadi et al. (Farhadi, Attarodi, Dabanloo, Mohandespoor, & Eslamizadeh, 2018). Statistical test, Analysis of Variance (ANOVA) was used to evaluate the extracted features. The model was evaluated on the MITDB and achieved accuracy of 93.6%. Another study based on SAE was proposed by (Yuan, Yan, Zhou, Bai, & Wang, 2016) for AF detection. The method was tested on AFDB, NSRDB, the MIT-BIH Long-Term, 24-h ambulatory ECG database with arrhythmia along with normal sinus rhythm. The model achieved accuracy, sensitivity and specificity of more than 96%. A study presented by Al Rahhal et al. (Mohamad Mahmoud Al Rahhal et al., 2018) also proposed SDAE network for learning feature representation from ECG data. Consequently, a softmax regression layer was added unto the SDAE network for automatic classification of Premature Ventricular Contractions (PVC). An overall accuracy of 98.6% was achieved on the INCART dataset. The summary of the Autoencoders for ECG signals is presented in Table 6.
-
VI.
Generative Adversarial Networks.
This section discusses the only paper that applied GANs-based model for ECG signal analysis. Based on this review, only one paper was found which applied GAN architecture for ECG arrhythmia classification (Z. Zhou, Zhai, & Tin, 2020) called GAN with Auxiliary Classifier for ECG (ACE-GAN). This model also addressed the problems faced by most of the DL architectures, such as CNNs on handling imbalance data and poor performance due to limited labeled ECG data. These problems have been investigated in the literature; however, the proposed methods still faced some challenges. For instance, labeling patient-specific heartbeats have been proposed to enhance classification performance. However, the systems were not fully automatic and there were too much experts’ involvements. To handle imbalance data, some studies have proposed cost-sensitive learning, random re-sampling and random under-sampling. Random re-sampling and cost-sensitive learning assign different weights to each class could lead to model over fitting and poor generalization ability (S. S. Xu et al., 2018). Random under-sampling involves selecting a sub-set of beats from the whole dataset. This also poses the problem of reducing the number of dataset required for effective training of CNN models (Kiranyaz et al., 2015). On the other hand, GAN may provide data augmentation and relieve class imbalance problem by learning to generate relevant conditional samples. The proposed ACE-GAN model used the generator network for data augmentation and the discriminator network was used for classification in a transfer learning manner. The model was evaluated on MITDB and achieved a competitive performance, with 99% accuracy on SVEB class and VEB class. The summary of the study is presented in Table 7.
-
VII.
Hybrid of DL Techniques.
In this section, we discuss papers that proposed the combination of different DL algorithms for effective classification/analysis based on ECG data. A 1D-CNN RestNet-34 in combination with LSTM was proposed for temporal relations feature extraction (Y.-J. Chen, Liu, Tseng, Hu, & Chen, 2019). The model is able to learn discriminative feature representation from ECG data and temporal relations using the LSTM. The model was tested with ECG data collected from the Taipei Veterans General Hospital Division of Cardiology ECG which yielded arrhythmia accuracy of 80%, sensitivity of 82% and specificity of 97%. A study presented by Chen et al. (C. Chen, Hua, Zhang, Liu, & Wen, 2020) proposed automated arrhythmia classification using CNN + LSTM model. The model was trained on MITDB and achieved 99.32% accuracy.
In another study for arrhythmia classification, a hybrid model of CNN and LSTM was proposed for feature extraction, combined with traditional extracted features (Chu, Wang, & Lu, 2019). They also used binary particle swarm optimization algorithm (BPSO) to select discriminating features which was fed into the SVM for the classification. This model was trained on MITDB and the performance shows 97.749% accuracy was achieved on the INCART database. Inter-patient ECG classification problem was investigated by Guo et al. (L. Guo, Sim, & Matuszewski, 2019). They proposed a pre-trained CNN model (DenseNet) and combined with RNN based model (GRU) for inter-patient ECG classification. The model was evaluated on the MIT-BIH Arrhythmia and Supraventricular Databases (SVDB). It achieved classification accuracy of 93.61% and 93.71% on SVEB and VEB classes, respectively. A research conducted by Ma et al. (Ma, Liu, & Chen, 2018) proposed a hybrid of CNN and RNN (CRNN) for ECG classification. The model was evaluated on MITDB and produced accuracy of 98.81%. A study conducted by Murugesan et al. (Murugesan et al., 2018) was conducted for arrhythmia classification. In the study three models, LSTM, CNN and hybrid of the LSTM and CNN (CLSTM) were proposed. Among the three models, CLSTM achieved the best performance with accuracy of 97.6% on the MITDB. However, the model was trained with imbalanced dataset. The authors proposed to train the model with noisy data to improve performance in real world.
A study presented by Oh et al. (Oh, Ng, San Tan, & Acharya, 2018) proposed a hybrid of CNN and LSTM for ECG arrhythmia diagnosis. The model used ECG segments of variable lengths from MITDB to classify different arrhythmias. However, the model required detection of the first R peak of the segment. Also, the model is not robust in detection of APB from normal ECG segment. More so, imbalanced data was used to evaluate the model. The proposed model produced 98.10% accuracy, 97.50% sensitivity and 98.70% specificity. Another study presented by Shi et al. (Shi, Qin, et al., 2020) proposed a novel system for automatic heartbeat classification using a combination of CNN and LSTM architectures. The novelty of this study is in the use of multiple input layers as opposed to most models that used only one input layer. They divided the heartbeat into three regions as the three input layers. These input layers were passed into the convolutional layers with different kernels strides and the resultant time series were inputted into LSTM and passed through fully connected layers. The output of this formation was concatenated with the fourth input layer, which is the RR interval features. The model was evaluated on MITDB for both class-oriented scheme and subject-oriented scheme. The accuracy achieved were 99.26% (class-oriented) and 94.20% (subject-oriented).
Detection of cardiac arrhythmia task was proposed by Swapna et al. (Swapna, Soman, & Vinayakumar, 2018). The authors proposed CNN and combination of CNN with RNN (RNN, LSTM and GRU). The hybrid of CNN and LSTM achieved the best accuracy of 83.4% when tested on the MITDB. A combination of CNN and bi-directional RNN called CBRNN was proposed by Wang et al. (E. K. Wang et al., 2019) for ECG diagnosis. The model was evaluated using the Chinese Cardiovascular Disease Database (CCDD) and 87.69% accuracy rate was achieved. A model based on the hybrid of convolutional AE (CAE) and LSTM for arrhythmia classification was proposed by Yildirim et al. (O. Yildirim, Baloglu, Tan, Ciaccio, & Acharya, 2019). The CAE was applied to compress large-sized ECG signals with minimum loss, thereby reducing storage cost and improving recognition performance. The LSTM performed the classification using the compressed ECG signals. The model was evaluated on the MITDB and achieved accuracy of 99.11% (using coded features) and 99.23% (using raw data).
Real-time detection of AF was investigated by Andersen et al. (Andersen et al., 2019) using the combination of CNN and bi-directional LSTM. The model was tested on three databases, MITDB, AFDB and NSRDB. 98.98% sensitivity and 96.95% specificity was achieved on AFDB. However, the automatic R-peak segmentation algorithm failed to detect R-peaks in multiple segments due to noisy ECG signals. Another study by Dang et al. (Dang, Sun, Zhang, Qi, et al., 2019) proposed a combination of deep CNN and bi-directional LSTM for AF classification. The model achieved training accuracy of 99.94% using the MITDB. And hybrid of CNN and LSTM model was proposed by Ivanovic et al. (Ivanovic, Atanasoski, Shvilkin, Hadzievski, & Maluckov, 2019) for detection of AF and Flutter task. The model was trained on the ECG data having 1097 30 seconds-long ECG recordings and achieved 88.28% average performance accuracy. A study by Li et al. (Dengao Li, Li, Zhao, & Bai, 2019) proposed an automatic heart failure staging of heart failure diseases. It presented a CNN-RNN based model for feature extraction, feature selection and classification. Feature extraction was done using the CNN structure. More so, clinical data was combined with the features extracted from CNN and fed into the RNN structure for classification. The dataset used to evaluate the model was collected from the chest pain centers (CPCs) of Shanxi Academy of Medical Sciences. The model achieved accuracy of 97.6%. Early detection of coronary artery diseases (CAD) is essential to avert the occurrence of MI and finally CHF. Another CNN-LSTM based model was proposed by Lih et al. (Lih et al., 2020) for the classification of MI, CAD and CHF. The proposed model was trained with data collected from PTBDB, Fantasia database, INCART AND BIDMC Congestive Heart Failure Databases. The model achieved the best performance of 98.51% accuracy. However, limited populations were used for CAD and CHF classes.
A study by Lui and Chow (Lui & Chow, 2018) proposed a CNN-LSTM based model with stacking decoding to improve classification accuracy. The proposed model was suitable for portable devices since it used single lead ECG recordings, which is also suitable for the portable and wearable medical and health care devices. The model was evaluated using the PTBDB and achieved 92.4% accuracy. A study presented by Picon et al. (Picon et al., 2019) put forward a study for the automatic detection of lethal Ventricular (LV) arrhythmia. The proposed model combines CNN and LSTM to automatically extract features for classification. Two sets of databases were used to evaluate the proposed model, one from public database, including three databases, namely MIT-BIH Malignant Ventricular Arrhythmia Database (VFDB), the CUDB and American Heart Association ECG Database (AHADB). The other database is from out of-hospital cardiac arrest (OHCA) database. The results of the evaluation yielded accuracy of 99.3% and 98.0% for both public database and OHCA database, respectively. However, there were difficulties in gathering annotated OHCA data. Detection of paroxysmal AF was investigated by Shashikumar et al. (Shashikumar, Shah, Clifford, & Nemati, 2018). Paroxysmal AF is another manifestation of AF that is often very difficult to be noticed. The authors proposed a CNN combined with Bidirectional RNN for the classification. The ECG data was first transformed into wavelet spectrograms and fed into CNN for feature extraction and passed into BRNN for capturing temporal patterns. The evaluation of the proposed model based on area under a curve (AUC) produced 0.94 using a private dataset collected from 2850 patients at the University of Virginia (UVA) Heart Station. Another CNN + LSTM based model was proposed by Verma and Agarwal (Verma & Agarwal, 2018) for AF detection task. The model was evaluated using the PhysioNet Challenge 2017 which yielded F1 score of 91.11% for AF classification.
The detection of CAD using manual method is challenging due to its low amplitude, an automatic system is essential to capture the abnormal ECG morphology for accurate identification. A study on CAD classification was put forward by Tan et al. (Tan et al., 2018). They proposed an automatic system by hybridizing CNN and LSTM for automatic features extraction and classification of CAD ECG morphology. The model achieved accuracy of 99.85%, sensitivity of 99.85% and specificity of 99.84%. However, the model was trained with imbalanced and limited data. More so, the model failed to achieve optimum diagnostic performance for subject specific data due to limited number of CAD.
Heart rate variability (HRV) derived from ECG signals have been found effective to detect diabetes. A study by Swapna et al. (Swapna, Kp, et al., 2018) put forward a novel study on the detection of diabetes. They proposed a CNN model and a combination of CNN and LSTM structures. They utilized a dataset collected from the diabetes and normal group of 20 people each to evaluate the models. The hybrid CNN-LSTM model achieved the best performance with 95.1% accuracy. In another study by Swapna et al. (Swapna, Vinayakumar, & Soman, 2018) they proposed a DL-based feature extraction model for detection of diabetes. The proposed model applied the combination of LSTM and CNN as feature extractor and employed SVM to classify HRV for binary diabetes classification. The model was tested on a private ECG dataset collected from the diabetes and normal group of 20 people each and achieved accuracy of 95.7%. However, these studies require larger dataset for training and testing to improve the generalization ability.
A study by Banluesombatkul et al. (Banluesombatkul, Rakthanmanon, & Wilaiprasitporn, 2018) proposed a 1D-CNN + LSTM based model for automatic feature extraction and temporal information extraction. A fully-connected DNN was employed for classification. However, the proposed model needs to be improved to detect different levels of OSA. The model was evaluated on the MrOS sleep study (Visit 1) database and achieved 79.45% accuracy. A study by Erdenebayar et al. (Erdenebayar, Kim, Park, Joo, & Lee, 2019) proposed six DL algorithms: DNN, 1D-CNN, 2D-CNN, RNN, LSTM and GRU for automatic detection of sleep apnea events. The GRU model achieved the best accuracy of 99.0%, 1D-CNN and GRU models achieved the best performance of 99.0% in terms of recall rates. However, the ECG signals were distorted by position changing, snoring, and cough during sleep. The summary of the studies that apply hybrid DL algorithms is presented in Table 8.
5.3.1.2 Biometric/Security domain
ECG-based biometric systems have drawn the attention of researchers in recent years. It is based on using the human heartbeat variability from ECG to identify and authenticate individuals. Though, ECG-based biometric models are mostly less accurate than the existing physiological traits such as fingerprint and iris traits. The ECG is considered effective because of its uniqueness and specificity (Muhammed & Aravinth, 2019). It is also considered more difficult to be compromised by spoofing attacks than the existing biological traits (Abdeldayem & Bourlai, 2019; Lynn et al., 2019). Another advantage of the ECG signal is that it is the only biometric feature that certifies the liveness of the subject (Chamatidis, Katsika, & Spathoulas, 2017; Hammad et al., 2019). The biometric system can be based on physiological characteristics, such as fingerprint (Bansal, Sehgal, & Bedi, 2011), iris (Bowyer, Hollingsworth, & Flynn, 2008) and hand veins (Sarkar, Alisherov, Kim, & Bhattacharyya, 2010) or behavioural traits, such as keystroke dynamics (Monrose & Rubin, 2000) and signature (Hafemann, Sabourin, & Oliveira, 2017). The ECG was found applicable as a biometric trait for human identification (Biel, Pettersson, Philipson, & Wide, 2001; Irvine, Israel, Wiederhold, & Wiederhold, 2003) and it falls under physiological characteristics (Bajare & Ingale, 2019). The features extracted from ECG signals were found to be unique to individual. ECG for Biometric systems was first introduced in 1977 by the US military (Forsen, Nelson, & Staron Jr, 1977).
A biometric system either identifies or authenticates an individual based on the unique features (Lynn et al., 2019; Pinto, Cardoso, & Lourenço, 2018). Identification process involves supplying input data and the model outputs the identity of the unknown subject. While in the authentication process, the model accepts or rejects claims of identity supplied with input data (Salloum & Kuo, 2017). However, the variability of ECG signals may present challenges in biometric systems. The inter-subject (uniqueness factors) and intra-subject (permanence factors) variability may be affected by many factors such as heart geometry, individual attribute, medication, cardiac condition, posture, age, emotion, fatigue, electrode characteristics and placement. These challenges remain open issues (Abdeldayem & Bourlai, 2019; Carreiras, Lourenço, Silva, Fred, & Ferreira, 2016; Pinto et al., 2018). These psycho-physiological factors should be considered during the acquisition of ECG for biometric systems (Pinto et al., 2018). Biometric identification based on ECG can be categorized into two major methods: fiducial and non-fiducial approaches (Chamatidis et al., 2017). The former involves describing the peaks, boundary and intervals of P, the QRS and T waves and the main three ECG waves. These methods depend on ECG signals segmentation and feature engineering that makes their generalization ability poor and inefficient (Bajare & Ingale, 2019). The latter instead of applying fiducial localization, which is computational cost, the complete signal processing is applied. These methods are based on waveform and morphology to extract features and usually process signal in the frequency domain (J. S. Kim, Kim, & Pan, 2020). A study in (Abdeldayem & Bourlai, 2019) identified hybrid ECG biometric-based identification that combines both fiducials and non-fiducials approaches. By utilizing the robust DL architectures, ECG data can be analyzed by passing through the hidden layers to extract unique features that can be utilized to identify and authenticate individuals. There are studies already in the literature that utilized DL architectures in ECG-based biometric systems and this is attracting the research community because of its unique benefits. The subsequent sub-sections discussed the various studies that used DL in ECG-biometric based systems.
-
I.
Deep Neural Networks for ECG Biometric authentication.
The DNNs application in ECG has been used in the domain of biometric systems to identify and authenticate humans. A DL network was proposed by Chamatidis et al. (Chamatidis et al. 2017) for human authentication. The model first converted the ECG signals using three transformations: Discrete Cosine Transform (DCT), Fourier Transform (FT), and Discrete Wavelet Transform (WT) and then fed into the DNN for authentication. However, the model performed poorly compared with the KNN classifier, MLP classifier, Radial basis function network (RBFN) classifier and Random Forest classifier, largely due to insufficient data for training. The authors proposed to incorporate cancelable mechanism to protect the system against security attacks. A study performed in 2015 by Page et al. (Page et al. 2015) proposed DNN model for user identification. The study utilized 90 subjects from ECG-ID database and obtained 99.54%, 99.85% and 99.96% performance of accuracy, sensitivity and specificity, respectively. However, the ECG signals were collected while the subjects are in resting state. Other events like during exercise, standing and so on, can be investigated. A DNN model was proposed by Wieclaw et al. (Wieclaw et al. 2017) for human identification. The study was evaluated on Lviv Biometric Data Set which contains 18 subjects’ recordings and produced accuracy of 88.97%. However, distortion of ECG signals and limited data affected the performance. Data augmentation based on GANs can be employed. The summary of the studies is presented in Table 9.
-
II.
Convolutional Neural Networks for ECG Biometric authentication.
There are CNN-based ECG-based biometric systems proposed in the literature for the sole purpose of identification and authentication problems. The ECG-based biometric systems have been considered more effective and hard to break by spoofing attacks. A 2D-CNN model was proposed for human identification (Abdeldayem & Bourlai, 2019). The model was tested on eight different databases and obtained the best average identification rate (IR) of 95.6% on the combined databases. However, the study suggested employing multi-session scenario to improve the model performance. A study by Bajare and Ingale (Bajare & Ingale, 2019) used 1D-CNN model for human identification. The model achieved accuracy of 96.93% using NSRDB and 100% using ECG-ID database. A study by Byeon et al. (Byeon, Pan, & Kwak, 2019) used three CNNs pre-trained models: AlexNet, GoogLeNet, and ResNet, for human identification. The ResNet showed high performance compared with the other pre-trained CNNs models on both PTBDB and Chosun University (CU-ECG) databases. However, the performance can be improved by employing ECG representations that are robust to noise. In another study (Byeon, Pan, & Kwak) proposed preconfigured CNN models using various time-frequency representations as inputs for human identification. The VGGNet, ResNet, DenseNet, and Xception models were proposed and Xception was found to be better than the compared algorithms. Also, the best among the time-frequency representations was mel frequency cepstrum coefficient (MFCC), was found better than spectrogram, log spectrogram, mel spectrogram, and scalogram. An Emsemble of Xception, ResNet, and DenseNet model were proposed (Byeon et al., 2020). The authors used spectrogram, melspectrogram, log-spectrogram, scalogram and MFCC to obtain time-frequency representations that were fed into the pre-trained CNNs models. Experiments result showed that the best accuracy of ensemble by time frequency representation was Xception with 99.05% using average of scores from log spectrogram to spectrogram. Also, the best accuracy of ensemble by opened CNN model was MFCC with 99.04% using average of scores from Xception to DenseNet.
A study by (Deshmane & Madhe, 2018) proposed 1D-CNN model for human identification and tested the model over four databases. The performance accuracies of 81.33% (MITDB), 96.95% (Fantasia database), 94.73% (NSRDB) and 92.85% (QT database) were obtained. Another pre-trained CNNs model, Inception-v3, was proposed by (P.-L. Hong et al., 2019) for identity recognition task. The model was tested on PTBDB and achieved an IR of 97.84%. However, the study could not be easily scaled. A study for personal identification task was proposed by Kim et al. (J. S. Kim et al., 2020). The model achieved the best accuracy of 99.2% on the NSRDB. However, the ECG signal acquisition under different activities was proposed to be integrated for better recognition ability. A parallel ensemble CNN model was proposed by Kim et al. (M.-G. Kim, Choi, & Pan, 2020) for user recognition. The authors proposed this model to deviate from the problem of over fitting when data obtained from different sources changes the user state is presented as registered data, which may lead to data generalization. Consequently, degrade the recognition performance of newly data. Therefore, the various ECG signals acquired from different states were passed into the ensemble networks and the output was fused into one database for re-training. The model achieved performance accuracy of 98.5%. However, the ECG signal was collected using only one device; this may affect the model’s generalizability. A study conducted in (Labati, Muñoz, Piuri, Sassi, & Scotti, 2019) proposed a CNN model for user recognition called deep-ECG. The model achieved 100% accuracy on PTBDB. However, to protect the system from security attacks, ECG-Deep binary features can be employed for template protection. User ECG recognition based on 1D-CNN was proposed by Lei et al. (Lei, Zhang, & Lu, 2016). The model was tested on PTBDB and achieved 99.33% accuracy using ECG data from 100 subjects. A novel study was proposed by (Y. Li, Pang, Wang, & Li, 2020). They proposed a CNN-based model called Cascaded CNN (F-CNN + M-CNN). The F-CNN model was used as feature extractor and M-CNN was used for the identification task. Both the two trained models were cascaded to form the cascaded CNN for final identification. The model performance is found to be better than the existing approaches with accuracy of 94.3% (3s, 18–40 subjects) and 97.1% (7s with 5 datasets). However, the model still suffers from intra-subject variability. A study by Zhang et al. (Q. Zhang, Zhou, & Zeng, 2017) proposed a 2D-CNN model for human identification. The study used Single-arm-ECG signal data acquired from 10 subjects and achieved IR of 98.4%. In another study, (Q. Zhang & Zhou, 2018) proposed 2D-CNN model for human identification. The model was tested on dataset acquired from single-arm-ECG and ear-ECG. Result shows it achieved IR of 98.4% and 91.1%, respectively. However, the ECG signals acquired from these studies were weak. A study by Zhang (Q. Zhang, 2018) proposed a CNN model for human identification. The experiment was carried out using NSRDB and an IR of 99.0% was achieved. The effect of more leads on the model performance was proposed to be investigated in the future.
Human authentication task was investigated using ID-CNN model proposed in (Ying Chen & Chen, 2018). The ECG signals of 50 healthy subjects were recorded by a BMD_Starter_Kit with a built-in BMD101 chip (NeuroSky Japan Inc.). The model yielded 2.0% EER over 12000 beats and the false acceptance rate (FAR) and false rejection rate (FRR) were less than 10.00%. A study proposed by Hammad et al. (Hammad et al., 2019) used CNN model for authentication. The features were first preprocessed and extracted using manual feature extraction and a 12-layer CNN was used for authentication. They introduced a new database called MWM-HIT database for training the model. The model was tested on the records collected from PTBDB, CYBHi and MITDB. On the first scenario EER of 5.97% and 12.69% was recorded on PTB and CYBHi, respectively. The second scenario produced EER of 1.63% and 4.47% on PTBDB and CYBHi, respectively. In another study by Hammad and Wang (Hammad & Wang, 2019) a pre-trained CNN based model was built, VGG-Net was used for feature extraction combined with QG-MSVM for authentication in a parallel score fusion of ECG and fingerprint. The model was tested on PTBDB and LivDet2015 database and accuracy of 99.99% was achieved. Another study presented by Hammad et al. (Hammad et al., 2018) combined VGG-Net and QG-MSVM for human identification. The pre-trained model (VGG-Net) was used as feature extractor and the features were passed into QG-MSVM for authentication. The model fused ECG and fingerprint features for effective biometric system. They also implemented a concealable method to seal the biometric template which enhanced the security of the system. The model was tested on different databases such as PTBDB, CYBHi database, FVC 2004 fingerprint database and LivDet2015. While the model showed better performance on both ECG and fingerprint databases, the overall performance on the multimodal database achieved EER of 0.14% (MDB1) and EER of 0.10% (MDB2). The performance can be improved by introducing different level of fusion. More so, the biometric features size can be reduced to speed up training.
Two CNN models were proposed by Hammad et al. (Hammad, Pławiak, Wang, & Acharya, 2020) for human authentication. The first model was a 1D-CNN and the second was a pre-trained model (ResNet) with attention mechanism called ResNet-Attention. The proposed ResNet-Attention model achieved better performance with accuracy of 98.85% and 99.27% on PTB and CYBHi, respectively. However, ECG template protection was not used. A study presented by (Ranjan, 2019) for user authentication investigated the permanence of the biometric and the impact on system accuracy on day-to-day variations in ECG. The system achieved EER of 2.0% (minimum EER of 0.9%) tested on PTBDB. However, the authors observed that the accuracy degrades due to single session enrollment as days pass by. Table 10 presents the summary of applications of CNN in ECG for biometric authentication.
-
III.
Recurrent Neural Networksfor ECG Biometric authentication.
RNN-based models were found in application for human identification and authentication. A study proposed by Salloum and Kuo (Salloum & Kuo, 2017) used RNN, GRU and LSTM models for identification/authentication. The LSTM model achieved the best performance with 100% accuracy on both MITDB and ECG-ID database. However, there were variations of ECG segmented window. Another study proposed by (Lynn et al., 2019) presented human identification task study using bidirectional GRU and bidirectional LSTM models. The models achieved accuracy performance of 98.55% (BGRU) and 96.4% (BLSTM). Table 11 presents the summary of the applications of RNN in biometric authentication using ECG signal.
-
IV.
Autoencodersfor ECG Biometric authentication.
An ECG-biometric based on AE was proposed by Eduardo et al. (Eduardo et al. 2017). The proposed method was tested on a private dataset collected from a local hospital using a Philips PageWriter Trim III device from 709 subjects. The model achieved a good performance. The performance of the proposed method can be improved by employing more robust preprocessing methods. Table 12 presents the summary.
-
V.
Hybrid of DL Techniques for ECG Biometric authentication.
From our review, only a single study was found to apply hybrid DL model for ECG signal analysis. A study presented by Muhammed and Aravinth (Muhammed & Aravinth, 2019) proposed an off-the-person biometric human authentication system using CNN and hybrid of CNN and LSTM models. The CNN + LSTM model achieved the best performance with accuracy of 84.63% on CYBHi dataset. However, noise and variability factors can affect the performance of the model. Table 13 presents the summary.
5.3.1.3 Driving domain
Due to the alarming rate of accidents occurring on the roads, systems based on ML have been proposed in the literature for automatic detection of driver’s state to alert the driver when needed. So that accidents could be averted. Accidents on the roads and air space are caused by many things and for so many reasons. For instance, an accident can occur as a result of fault developed by the vehicle or as a result of collision with another vehicle or as a result of a human being or an animal mistakenly barge in front of a running vehicle and so on. On another angle, there are accidents that are caused by the driver’s inattention, distraction, stressed or drowsiness and so on. Driver drowsiness detection systems proposed may be categorized into three approaches (Abbas, 2020; Jabbar et al., 2018). The first approach is based on driving patterns. This is based on vehicle characteristics, road conditions, and driving skills. The second approach is based on physiological sensors data, which includes: ECG, EOG, and EEG data. These techniques performances are largely dependent on attaching sensors on the driver’s body which is intrusive. The third approaches used computer vision to detect driver’s activities such as yawning duration, head movement, gaze or facial expression, and eye closure. The best ways for correct and relatively accurate detection depend on human drivers physiological signals such as brain waves and heartbeats (Abbas, 2020).
These systems utilize human heartbeat (ECG signals) to detect certain conditions of a driver thereby triggering an alarm for necessary actions to be taken to prevent accidents. Various methodological techniques for the detection and prediction of driver fatigue have been developed through advanced image processing, machines learning, and computational intelligence algorithms. The real-time, accurate driver fatigue and drowsiness detection can reduce the rates of accidents (Bhardwaj, Natrajan, & Balasubramanian, 2018). Other applications based on classification of pilots’ mental state have been proposed (Han, Kwak, Oh, & Lee, 2020). These methods have been designed to detect pilot’s mental status such as distraction, workload and fatigue, which could help prevent possible airplane crash due to deteriorated cognitive state of pilots. The following sub-sections, discussed the papers that applied DL in ECG signals with the aim to reduce the rate of accidents while driving.
-
I.
Deep Neural Networks.
In driving application, stress detection has been used to detect driver’s stress based on the driver’s ECG signals using DL. A study by Cho et al. (Cho, Park, Dong, & Youn, 2019) proposed a DNN model to detect stress using driving data and mental data. They recorded a performance accuracy of 90.19%. The data acquired from this study was from specific stressful tasks. This can be improved for daily stressful monitoring. The model can also be used to detect anxiety which is a potential stressor to so many health challenges like high blood pressure etc. Table 14 presents the summary.
-
II.
Convolutional Neural Networks (CNNs).
The CNN-based models were found applicable in driving domain for inattention identification, stress detection and workload prediction. A study for inattention identification was presented by Taherisadr et al. (Taherisadr et al., 2018) to provide an early distraction detection system that could help prevent accidents on the roads while driving. They first extracted Melfrequency spectral coefficients (MFSC) representation of raw ECG signals and fed the 2D spectrogram images into the proposed deep CNN and achieved accuracy of about 95.51% which was better than using time frequency (TF) representation. The model was tested with ECG signals acquired from naturalistic setting driving of 10 persons. The limited population used can be increased to get more data to train the model. In another study, Pilot workload prediction was proposed (Xi et al., 2019). Two visual representations: spectrograms and scalograms were used on the acquired ECG. And a pre-trained CNN model, AlexNet was used as “off-the-shelf” feature extractor and the well-known linear classifier, SVM was used for the prediction. Compared with the finetuning Deep CNN, AlexNet + SVM achieved the best performance with accuracy of 51.35% (scalograms + AlexNet-SVM). However, the dataset was collected from 2 qualified test pilots performing a target tracking task on the National Research Council of Canada’s (NRC) Bell 205 helicopter which is very limited for a real setting. Table 15 presents the summary.
-
III.
Autoencoders.
From our review, a single study was found to have proposed AE-based model to model ECG signals. The study proposed a driver fatigue classification using deep AE (Bhardwaj et al. 2018). Data was collected using 10 subjects recorded during driving simulator and achieved accuracy of 96.6%. A Multimodal system integrating biosignals (ECG, EEG and EMG) was suggested for robust driver fatigue system. Table 16 presents the summary.
-
IV.
Hybrid of DL Techniques.
The Driver’s Drowsiness Detection (DDD) system was proposed by Abbas (Abbas, 2020) called “HybridFatigue”. The author proposed a transfer learning approach by employing CNN and DBN to detect driver’s fatigue from hybrid features in real time. The model utilized both the visual and physiological signal information through the use of a camera and ECG signals respectively, to obtain features for training. The proposed model was evaluated using three online databases: Columbia gaze dataset (CAVE-DB), DROZY, and closed eye wild (CEW) datasets. The pre-trained CNN + DBN achieved the best performance with accuracy of 94.50% compared with the basic CNN model. The performance of the model can be improved by integrating a 3 multi-scam approach to improve recognition. A study conducted by Rastgoo et al.(Rastgoo et al., 2019) proposed the detection and classification of driver stress level using a multimodal fusion model based on hybrid of CNN and LSTM structures. The proposed model was employed to learn highly correlated representation from a multimodal fusion of ECG, vehicle data and contextual data. A private dataset collected from an advanced driving simulator was used for evaluation, the model achieved accuracy of 92.8%, sensitivity of 94.13%, specificity of 97.37% and precision of 95.00%. However, the study utilized very limited population. Table 17 presents the summary of the studies.
6 General analysis and discussions
This section presents discussion and analysis of the applications of DL in ECG signals from different domains of application as reviewed in the previous sections. We reviewed and analyzed primary studies based on domain of applications, DL models, application area, application tasks, dataset sources and, processing methods.
6.1 Discussion of the Deep Learning model
In this sub-section, we discussed the summary of the studies that applied DL to model ECG data. The summary presented is based on the DL applied.
RQ2: What are the DL techniques that have been applied for ECG signal analysis ?
Question 2 seeks to find out the DL techniques/models that have been used for ECG signal analysis, in the studies included for this review. Based on the review, different DL algorithms have been proposed in the literature for ECG modeling based in different domains (see Table 2–17).
Figure 18 shows the number of DL algorithms proposed for ECG data modeling as reviewed in this study. The CNN based models show consistency in application across the three domains having the highest number of papers. This may be because the CNNs models are suitable for ECG images classification since ID ECG signals can be transformed into 2D image representations for classification. In addition, it may be attributed to the success of the CNN-based models such as AlexNet, ResNet, VGG-Net and googLeNet for image classification. The RNN-based models so far have found applications only in medical/healthcare and biometric/Security domain, no paper was found to apply RNN-based model in driving domain. Also, the RBM-based models are only applied in medical/Healthcare domain. The AE-based models found applications in all the three domains. However, only a single paper was found in biometric and driving domains, respectively. The GAN-based models have not been given much attention for ECG signal modeling. Based on this review, only a single study was proposed for ECG arrhythmia classification (Z. Zhou et al., 2020). However, GAN has potential for solving imbalance dataset (Pyakillya et al., 2017; Shaker et al., 2020; Z. Zhou et al., 2020). We also identified studies that combined different DL techniques to model ECG signals, applied across all the presented domains. As revealed in this review, the medical/healthcare domain has received a considerable number of research based on hybrid DL algorithms in ECG. More so, the hybrid of CNN-based and RNNs-based models found the highest applications for modeling ECG signal, especially the combination of CNN and LSTM-based models. Figure 18 shows attested to that. Studies based on DL for ECG data in driving domain are minimal. New and novel research are needed to help curtail the problem of accidents on the roads. Overall, it is evidenced that DL techniques have found application in ECG data and the rate of research keep increasing as the DL popularity keep increasing. Therefore, more research are needed to investigate the potentials of using DL to model ECG data and the existing studies can be improved for better performance.
6.2 Discussion of the application area
RQ3: In what application areas the proposed DL models were presented ?
Question 3 seeks to find out in what application areas are the proposed DL models presented.
Figure 19 shows the taxonomy of application areas where the proposed DL models were presented for ECG data analysis. The application areas are based on the domains found to be applied in this review study. The application areas found in the driving domain are categorized into workload/stress, drowsiness, fatigue and distraction/inattention classification (see Tables 14, 15, 16 and 17). Yet, there are only few papers found in these application areas. The biometric/security domain application area is either identification or authentication classification. As shown from Tables 9, 10, 11, 12 and 13, the identification area received much attention than the authentication area. Nevertheless, there was a single study found to have been presented for the two areas, that is, identification and authentication (Salloum and Kuo 2017). In the medical/healthcare domain, the application areas are categorized into corona artery disease, heartbeat disease (these included specific heartbeat diseases, such as AF, MI, CHF, etc.), general arrhythmia diagnosis, sleep stage, stress and diabetes. Based on the presentations in Tables 2, 3, 4, 5, 6, 7, 8, 9, 10, 11 and 12, general arrhythmia diagnosis received the highest number of papers, followed by heartbeat disease and followed by sleep stage. Only few papers were found for corona artery disease, stress and diabetes classification areas.
6.3 Discussion of the classification Task
RQ4: What are the application tasks performed by the proposed DL models ?
Question 4 seeks to investigate the tasks performed by the DL models. Figure 20 depicts the different application tasks performed by the DL models as surveyed in this study. Each of the domains: medical/healthcare, biometric, and driving have different and unique tasks applicable to each. A biometric system either identifies or authenticates an individual based on the unique features (Lynn et al. 2019; Pinto et al. 2018). Therefore, the ECG biometric systems perform only binary classification, which either accepts or rejects an individual based on the physiological or behavioural patterns of a person. From the presentations in Tables 9, 10, 11, 12 and 13, it can be deduced that human identification or recognition task received high applicability than the authentication task. The DL model’s application tasks proposed for driving based systems have been found to include driver’s drowsiness, stress level, pilot’s workload, driver’s fatigue, driver’s distraction and inattention detection.
All these tasks have not been investigated fully, only a few papers found to have implemented the tasks. The application tasks performed by DL models for medical diagnosis include AF and flutter detection, PAF detection, CAD classification, MI detection, CHF detection, diabetes detection lethal ventricular classification, OSA detection, fetal ECG classification, ventricular arrhythmia identification, stress classification and general diagnostic arrhythmia. Based on our survey, most of the studies were for heartbeat signal and diseases classification with AF classification being the highest investigated heartbeat diseases. The AF has been reported as the most investigated CVDs (Xia et al. 2018). Also, classification is considered one of the most machine learning problem used in healthcare and bioinformatics, notably for arrhythmia detection (Sannino and De Pietro 2018). Figure 20 shows the taxonomy of the application tasks performed based on the three domains found in the literature review.
6.4 Discussion of the dataset sources
RQ5: What are the sources of datasets utilized in the proposed deep learning models ?
Question 5 seeks to present the different sources of the dataset utilized in different studies for ECG modeling using DL. Based on our review, we observed three different types of dataset sources used: private, public and hybrid. The private datasets were acquired by the authors. This is mostly when there is no publicly available dataset for the task they intend to model. The public datasets are publicly accessible on the internet. The hybrid datasets are datasets that are formed from the combination of both the private and public dataset (Rim et al., 2020). However, in this review, we categorized these datasets based on the domain of collection, that is, medical/healthcare, biometric/security and driving domains. Also, we presented the number of datasets based on the number of usages. Figure 21 shows a bar chart depicting the number of dataset sources. The bar representing “Others”, are dataset sources that were not able to be represented singly (they are from the private dataset sources).
From Fig. 21 and in the category of medical/healthcare domain, the first 5 dataset sources mostly used by researchers for modeling ECG includes: MITDBFootnote 12 (60), PTBDBFootnote 13 (12), AFDBFootnote 14 (12), Physionet/CinC Challenge 2017Footnote 15 (12) and NSRDBFootnote 16 (8). In the domain of biometric/security, the first top 5 dataset sources mostly used are: PTBDB (12), MITDB (6), NSRDB (6), ECG-IDDBFootnote 17 (5) and CYBHi-DBFootnote 18 (4). In the category of driving domain, there are mainly three dataset sources: CAVE-DBFootnote 19, DROZZYFootnote 20 and CEW-DBFootnote 21, with single usage. Other dataset sources are private sources and are grouped in the “Others” category.
6.5 Discussion of the Preprocessing Method
RQ6a: What ECG preprocessing methods and training architectures were used by the proposed DL model?
Research question 6a seeks to investigate the ECG preprocessing methods and training architectures used by the DL to model ECG. After the acquisition of data from the ECG devices, there are 4 stages that can be employed to analyze input data for classification purposes (Jeon et al., 2019; S. Zhou & Tan, 2020). First, preprocessing stage: this is where the input data is prepared by removing baseline wander, powerline interference and noises in the data. This improves the quality of the raw data and consequently enhances classification accuracy (Wu et al., 2016) (Serhani, T El Kassabi, Ismail, & Nujum Navaz, 2020). Second, heartbeat segmentation: this involves splitting the ECG signals into segments or partitions following the detection of QRS peaks. The QRS complex has important features relevant and helpful in separating different types of heartbeat (Jun et al., 2016). The third stage is feature learning: this is the stage where features are extracted and selected for automatic classification. The performance of the classification models is significantly dependent on the quality of the features extracted (Hsieh et al., 2020; Jeon et al., 2019; Wu et al., 2016). And in most times, if relevant and important features are not extracted, it can affect even the best classifier’s performance (Luo et al., 2017). The final stage involves the classification or detection or prediction of the desired classes.
During the ECG data acquisition using different ECG machines and scenarios, it is often accompanied with some signal distortions or noise contamination. This noise and artifacts are caused by so many factors including contraction of the muscles, poor electrode contact, snoring, coughing, respiratory movements and even the presence of other external devices (Isin and Ozdalili 2017). These distortions can be categorized into three namely Baseline Wander, Power line Interference (which introduces 50 to 60 Hz from the normal 0.5 to 80 Hz) and Electromyography Noise (Deshmane and Madhe 2018; Zhang 2006). These deviations from the normal ECG signals can drastically lower the performance of a model. Therefore, there is need to clean and remove these distortions, hence, the introduction of the pre-processing methods to process these signals before feeding them into the network for classification.
Based on our review, we observed that some studies applied pre-processing method over the raw ECG signals while some studies did not consider pre-processing at all. The studies that did not consider pre-processing method trained their models using the raw ECG signals. On the other hand, there are some studies that applied time frequency analysis techniques and transform the original 1D ECG signal to 2D ECG image representations and these representations are fed into the models for training. We also indicated the studies that applied segmentation method, partitioned the ECG signals into segments of different lengths and seconds following the preprocessing stage (or without the preprocessing stage) and the segments are fed into the models for learning. This segmentation follows the signal peaks detection, such as QRS complex.
Tables 18 and 19, and Table 20 present different pre-processing methods, signal peaks detections, segmentation and time frequency signals techniques applied on the raw ECG signals before inputting the processed signals into the models for training. To remove noise, baseline wander and power line interference, preprocessing using filtering methods were used such as median filter, 12-order low-pass filter, 6-order Butterworth bandpass filter, band pass filter, notch filter, bandpass second-order Butterworth filter, fourth order Butterworth band pass filter, finite impulse response filter, average filter, Daubechies wavelet 6 filter, Daubechies wavelet (db8), 150th-order bandpass Finite Impulse Response (FIR) filter, high pass filter, band reject filter, Savitzky Golay filter, two median filters, 12-tap low-pass FIR filter and so on. Also, some studies employed sampling techniques such as up-sampling, down-sampling and resampling on the ECG signals. More so, normalization methods have been used by some studies on the ECG signals. Normalization refers to scaling of the ECG signal samples to the same level with another sample (Pandey and Janghel 2020).
In our view, Z-score normalization is the most widely used normalization method based on evidence from the review. In the studies that used segmentation, signal peaks detection such as R and QRS peaks were mostly employed with Pan and Tompkins algorithm as the most utilized method for the detection of QRS components. Pan and Tompkins algorithm process ECG signals using filter, derivation, squaring, averaging and peak detecting (Debnath et al. 2018). It is considered one of the popularly used method in detecting QRS complex (Wieclaw et al. 2017). On the other hand, different frequency analysis techniques have been used by some studies to transform the 1D ECG signals to 2D ECG image representations, such as STFT, discrete wavelet transforms (DWT), FT, WT and CWT techniques. The popularly used methods for the frequency analysis of signals are DCT, CWT, and DWT (Byeon and Kwak 2019).
6.6 Discussion of the Learning Architecture
In a review paper presented by (Rim et al., 2020), they identified two approaches that input data can be learned.
First approach: feature extraction is performed on the input data and the features are fed into the network for classification.
Second approach: The raw data is inputted directly into the network for automatic classification.
There are three ways DL has been involved in the training. The DL have been used as feature extractors or as classifiers (first approach) or as automatic classifiers or end-to-end learning (second approach). The following sections discussed these training architectures in detail.
-
I.
Deep Learning as Feature Extractor.
There are studies that presented architectures based on DL as feature extractor and traditional method as classifier. These architectures aim to deviate from the cumbersome labeling of dataset using hand-crafted methods. Thereby employ the DL is trained using the dataset in an unsupervised learning manner. The outputs of the networks are usually fed into the traditional statistical and ML methods such as SVM, MPL, KNN, binary trees and ELM. Table 18 presents the summary of the DL models.
-
II.
Deep Learning as a Classifier.
There are some studies that proposed models using traditional method as feature extractor and DL as the classifier. This is with the aim to enhance classification accuracy by transforming raw data into feature data that has better discriminated characteristics. And these features are fed into the DL for automatic classification. In view of the fact that ECG signals are ID signals, some studies applied different transformation methods to convert these signals into 2D images. These new representations are then fed into the DL networks for classification. Table 19 presents the summary of the proposed models that applied this approach.
-
III.
End-to-End Learning.
In some approaches, the raw ECG data is entered directly into the network for automatic classification. In this approach, a single DL or hybrid of the DL, have been used for a more robust classification. This approach is used to demonstrate the robustness of the DL to automatically extract features, perform feature selection and classification. For instance, combinations of CNNs and RNNs-based models have been proposed (Andersen et al., 2019; Banluesombatkul et al., 2018; C. Chen et al., 2020; Chu et al., 2019; L. Guo et al., 2019). These studies used CNN which is basically developed for feature extraction to extract salient features in the dataset and used LSTM for temporal variance analysis of the data during modelling. This way, the model is able to explore the characteristics of the data for a better performance. Table 20 presents the summary of studies that applied this learning approach.
Figure 23 depicts the DL role in the learning of the proposed models as reviewed in this study. The roles of DL architectures have been categorized into feature extractor, classifier and end-to-end learning. In the category of DL as feature extractor, CNN-based models received the highest applications (15 papers, 10%). Followed by RNN-based models with 2 papers, then RBM-based models and hybrid models had single paper each. The AEs and GAN-based models were not found applicable in this category. In the category of DL as classifier, DNN, CNN and RNN-based models had 3 papers each that used DL as classifier. While RBMs and AEs each had single paper each. The GAN and hybrid-based networks were not found applicable in this category. Lastly, the end-to-end learning category found applications across all the discussed DL architectures. The CNN-based models had the highest application with 64 papers (about 42% ). The hybrid models received the second highest number of papers (25 papers), followed by RNN-based models (11 papers), and followed by DNN-based models with 9 papers. The AE-based models had 7 papers. The RBM and GAN-based models had 3 and 1 papers each respectively. Based on our review, only a single paper used GAN to model ECG data. Also, it can be deduced that application of DL in ECG is gaining attention with end-to-end learning models.
7 Challenges and future research directions
This section highlights the challenges considering the reviewed papers and discusses potential opportunities for further research. We categorized the challenges into domain, model, application task, dataset and DL architecture. Consequently, opportunities are highlighted for improvements.
7.1 Domain Challenges
Based on our review, we categorized the application of DL in ECG into three domains: medical/healthcare domain, biometric/security domain and driving domain. Even though, biometric domain applications can be considered as healthcare applications, because most of the applications in biometric using ECG are healthcare applications. These domains have received a lot of interesting research over the years. However, the nature of life does not guarantee that a patient can be present at the hospital for diagnosis all the time. It is also possible that some abnormalities surface at irregular times like cardiac arrest, when the patient is away from the hospital or under a physician’s examination. Therefore, the development of healthcare applications that monitor a patient in real time, alerting physicians in real-time to provide better assistance at the right time is highly necessary. Health and medical care applications are considered as one of the most fascinating applications that can fully benefit from the IoTs deployment. Although there are studies that have implemented DL to process ECG signals for edge computing and IoTs deployment (Azimi et al., 2018; Konan & Patel, 2018) (Farahani, Barzegari, & Aliee, 2019) (Granados, Chu, Zou, & Zheng, 2019). However, requirements of latency, low power and knowledge extraction from the large volume of physiological data are challenge for real time applications. Edge computing environment, modern AI techniques combined with 5G speeds would best meet the necessary requirements for the latency, accuracy and energy efficiency during real-time collection and analysis of health data (Hartmann, Farooq, & Imran, 2019). Therefore, applications of DL on ECG based systems can be developed and deployed on the cloud computing-based-environments, such as edge computing, software-defined computing, fog-computing, mobile edge computing, serverless computing and volunteer computing to enable ubiquitous and remote health monitoring(Buyya et al., 2018; Varghese & Buyya, 2018).
7.2 Model Challenges
Figure 18 presented the different DL models used to model ECG data for different tasks analysis. It is revealed that DNNs, CNNs, RNNs, RBMs, AEs, GANs based models and the hybrid have been proposed in the literature for ECG data analysis. There are different architectures of CNN including AlexNet (Krizhevsky et al. 2012), ZF Net (Zeiler and Fergus 2014) GoogLeNet (Szegedy et al. 2015), VGGNet (Simonyan and Zisserman 2014), ResNet (He et al. 2016), SqueezeNet (Iandola et al. 2016), DenseNet (Huang et al. 2017), Inception V3 (Szegedy et al. 2015), Xception (Chollet 2017), LeNet-5 (LeCun et al. 1998) and their variants. But as shown in this review, CNN architectures such as ZF Net, SqueezeNet and Lenet-5 may have not been giving serious attention for ECG modeling. It was found that AlexNet, ResNet and GoogLeNet have been used the most for ECG modeling. Also, there are different architectures of RNN, which include LSTM (Hochreiter and Schmidhuber 1997), GRU and bidirectional RNNs. It is found from the review that the LSTM models have been mostly used for ECG signal analysis. The DBN and DBM are both considered the “Boltzmann family” (Voulodimos et al. 2018). However, DBM models were not found applicable for ECG data analysis to best of our knowledge. There are also different architectures of AE (Zhai et al. 2018), which include Sparse AEs (SAEs), Denoising AEs (DAEs), Convolutional Autoencoder (CAEs), Variational AutoEncoder (VAE), Adversarial AutoEncoder (AAE) and so on. Only SAEs and DAEs have been found to be applied in ECG signal analysis. Also, there are studies that combined different DL structures such as CNNs and RNNs networks. Based on our review, the combination of CNN architectures and LSTM have been researched the most and have produced better performance compared to the single DL and other hybrid DL models.
The DL models have proven superior over the conventional ML models with better classification performances. However, the “black box” nature of DL models posed the challenge of interpretability. It is important that a more accurate diagnosis is achieved but it would add less value if it cannot be easily explained, especially in medical related diagnosis. In a bit to maintain interpretability in the DL models, some studies have combined deep features with traditional features of the models (Chu et al. 2019). A good discussion on the problem of DL interpretability can be found in (Miotto et al. 2018) (Hong et al. 2020). However, attempts are being made by researchers to provide interpretable and transparent models. A new computing paradigm called eXplainable Artificial Intelligence (XAI) (Doran, Schulz & Besold, 2017; Lipton, 2017; Mathews, 2019; Millier, 2018) is gaining attention by AI research community which tries to provide reasons or explain decisions and actions in a model, making the models transparent while providing a good accuracy. To build systems that can be trusted by humans, especially, for medical diagnosis, future ML and DL models must be explainable, explaining the causes to the effects or actions taken by the systems (Doran, Schulz & Besold, 2017). Another new paradigm based on Active Learning (AL) that applies XAI is being proposed in the literature, called eXplainable Active Learning (XAL) (Ghai et al., 2017). This paradigm was proposed to teach ML models, by having a model selectively query the machine teacher, and at the same time allows the teacher to understand the model’s reasoning and adjust their input, resembling a “teaching” experience. The XAI and XAL paradigms can be suitably applied on ECG data for various analyses and applications.
In the field of biomedical signal processing, the accuracy of a model’s classification depends largely on the quality of the signals and features extracted for classification. ECG feature extraction and classification are very critical in diagnosing CVDs. Dictionary learning algorithms have been successfully used in compression of ECG or noise elimination and feature extraction to effectively classify ECG especially, for medical diagnosis (Lee, Luan & Chou, 2014; Majumdar & Ward, 2017; Liu et al., 2016; Balouchestani, Sugavaneswaran & Krishnan, 2014; Mathews, 2017; Ceylan, 2018). More studies that leverage dictionary learning and DL for ECG signal classification are potentials for investigation in the future.
7.3 Application Task Challenges
There are different tasks executed by the DL models discussed in this review. We considered models based on their respective task. The models in each of the domain discussed have different targets to achieve during modelling. Figure 20 presented the taxonomy of these tasks and categorized based on domains. For example, in biometric ECG based systems, the model performed authentication for a user, to either allowed access or denied (see Tables 9, 10, 11, 12 and 13). However, there are still challenges of inter-subject and intra- subject ECG variability in biometric ECG based systems. The inter-subjects variability deals with the uniqueness issues and intra-subject deals with the permanence issues, that affect the system by factors such as heart geometry, individual attribute, medication, cardiac condition, posture, emotion, age and fatigue, electrode characteristics and placement (Abdeldayem and Bourlai 2019; Carreiras et al. 2016; Pinto et al. 2018).
These challenges remain open issues for ECG-biometric based systems. Also, some studies performed classification task on whether an instance exist or not. For example (Swapna, Kp, et al., 2018; Swapna, Soman, et al., 2018) classified ECG signals as diabetes or non-diabetes and normal or arrhythmia, respectively. (Shashikumar et al. 2018) classified as PAF or normal, (Yuan et al. 2016) classified as AF or normal, (Wang et al. 2019) classified ECG as normal or abnormal, (Vullings 2019) performed prenatal detection of CHD as CHD or Healthy, (Tan et al. 2018) classified CAD as normal or CAD, (Mostafa et al. 2017) classified apnea as apnea or not apnea (Taherisadr et al. 2018) detected driver’s inattention as distracted or not distracted and so on. Other studies classified output into groups or level of instances. For instance, (Abbas 2020) performed driver’s drowsiness detection as drowsy, sleepy and normal, (Luo et al. 2017) classified ECG beats into 5 classes. A study by (Urtnasan et al. 2018) performed apnea/hyponea detection as Normal, Hypopnea, Apnea, etc.
7.4 Dataset challenges
Based on our review, we identified more than 40 datasets that have been used in modelling DL to analyze ECG signals for different classification tasks. It can be deduced that MITDB, PTBDB, AFDB, physionet/CinC Chanllenge 2017, NSRDB, ECG-IDDB and CYBHi-DB have received the highest number of usage (Table 21). All these datasets are available online and can be accessed via the internet. However, the number of private datasets indicates their non-availability on the internet for public use. This had forced the authors to acquire it themselves with additional resources. More so, the commonly used dataset is MITDB (Moody et al. 2001), which is over 40 years and contains only records from 47 subjects and is grossly imbalanced. In addition, this database is a single session database, which is not adequate for experiments on biometrics (Hammad et al. 2019). The recent datasets used are PhysioNet/CinC Challenge 2017 (Clifford et al. 2017) and 2018 China Physiological Signal Challenge (Liu et al. 2018), but these database were recorded on short-term durations.
DL are data-driven, DL require large scale data for training (Qayyum et al. 2019). Tables 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16 and 17 presented limitations for each of the studies. Generally, it shows that the datasets used to train the models were not large enough to give a better generalization. In a bit to address the problem of imbalanced and small dataset in most of the existing databases, studies have been proposed such as data augmentation (Giannakakis et al. 2019) Hammad et al. 2018; Hammad and Wang 2019; D. Li et al. 2019) (Shaker et al. 2020) and transfer learning (Pan and Yang 2009; Weiss et al. 2016). Techniques such as SMOTE (Pandey and Janghel 2019) (Chu et al. 2019) and GAN (Shaker et al. 2020), were used to solve the problem of imbalance dataset.
Therefore, it is recommended that large scale datasets should be produced for all the domains discussed in this paper. These datasets should be made available online to encourage researches and ease the data acquisition process.
7.5 Training Architecture Challenges
Based on our review and evident is (Rim et al. 2020), there are three different approaches of learning architectures presented in the literature that used DL to model physiological signals. In this review, we categorized the proposed DL models for ECG signal analysis based on their role in the learning architecture (Fig. 22).
RQ6b: Which of the training architectures produced the best performance?
Question 6b seeks to find out the best architecture among the three presented in Tables 18, 19 and 20. However, we could not identify a study that implemented the three learning architectures using the same dataset and/or pre-processing method for modelling ECG signal. As such, the best architecture could not be ascertained. Therefore, novel research can be conducted to close this gab.
Other inherent challenges like security, latency, heat consumption, speed and efficiency should be considered when designing models, especially in medical and healthcare applications. The DL is application independent (Alom et al., 2019). The application of DL in technology have been reported in many areas, such as computer vision, natural language processing, finance, remote sensing, transportation, education, marketing and advertising, etc (Bote-Curiel et al., 2019). However, further improvements on these systems are necessary to achieve high acceptability of its application in the real-world settings.
8 Conclusions
This study presented a systematic literature review on the applications of DL to model ECG data from different application domains. The study revealed the superiority of DL models over the traditional ML methods on modeling ECG data. The paper discussed biometric ECG based systems and analyzed empirical studies that adopted DL for ECG signal processing based on the domain of application, application area and application task, the DL model, dataset source, preprocessing method, and training architectures. The study showed an increasing interest in the application of DL in ECG from the last decade and justifiably so, especially for medical and healthcare applications. This is expected to grow as the DL architectures become more popular. The DL application in modeling ECG data has proven to produce state-of-the-art performances and even more accurate performances than the experienced Cardiologists. However, apart from the well-known challenges of limited dataset and computational cost required by the DL applications, other challenges, such as security of applications, latency, low power and knowledge extraction from the large volume of physiological data for real time applications and the “black box” nature of DL models were also highlighted to enable room for future development. This study can serve as a benchmark material for new researchers to further improve the performance of the existing DL in modelling ECG signal.
Notes
References
Abbas Q (2020) HybridFatigue: A Real-time Driver Drowsiness Detection using Hybrid Features and Transfer Learning. (IJACSA) Int J Adv Comput Sci Appl 11(1):585–593
Abdeldayem SS, Bourlai T (2019) A Novel Approach for ECG-Based Human Identification Using Spectral Correlation and Deep Learning. IEEE Trans Biometrics Behav Identity Sci 2(1):1–14
Acharya UR, Fujita H, Lih OS, Adam M, Tan JH, Chua CK (2017) Automated detection of coronary artery disease using different durations of ECG segments with convolutional neural network. Knowl Based Syst 132:62–71
Acharya UR, Fujita H, Lih OS, Hagiwara Y, Tan JH, Adam M (2017) Automated detection of arrhythmias using different intervals of tachycardia ECG segments with convolutional neural network. Inf Sci 405:81–90
Acharya UR, Fujita H, Oh SL, Hagiwara Y, Tan JH, Adam M (2017) Application of deep convolutional neural network for automated detection of myocardial infarction using ECG signals. Inf Sci 415:190–198
Acharya UR, Fujita H, Oh SL, Hagiwara Y, Tan JH, Adam M, San Tan R (2019) Deep convolutional neural network for the automated diagnosis of congestive heart failure using ECG signals. Appl Intell 49(1):16–27
Acharya UR, Fujita H, Oh SL, Raghavendra U, Tan JH, Adam M, Hagiwara Y (2018) Automated identification of shockable and non-shockable life-threatening ventricular arrhythmias using convolutional neural network. Future Generation Computer Systems 79:952–959
Acharya UR, Oh SL, Hagiwara Y, Tan JH, Adam M, Gertych A, San Tan R (2017) A deep convolutional neural network model to classify heartbeats. Comput Biol Med 89:389–396
Ahmed I, Qasim F, Ali MNY (2019) Analysis & Automated Prediction of Myocardial Infarction Disease Using Optimized Deep Learning Architecture. Paper presented at the Proceedings of the 2019 7th International Conference on Computer and Communications Management
Al-Huseiny MS, Abbas NK, Sajit AS (2020) Diagnosis of arrhythmia based on ECG analysis using CNN. Bull Electr Eng Inf 9(3):988–995
Al Rahhal MM, Ajlan A, Bazi N, Hichri YAl, Rabczuk T (2018) Automatic premature ventricular contractions detection for multi-lead electrocardiogram signal. Paper presented at the 2018 IEEE International Conference on Electro/Information Technology (EIT)
Al Rahhal MM, Bazi Y, Zuair A, Othman M, BenJdira B (2018) Convolutional neural networks for electrocardiogram classification. J Med Biol Eng 38(6):1014–1025
Al Rahhal MM, Bazi Y, AlHichri H, Alajlan N, Melgani F, Yager RR (2016) Deep learning approach for active classification of electrocardiogram signals. Inf Sci 345:340–354
Alghamdi A, Hammad M, Ugail H, Abdel-Raheem A, Muhammad K, Khalifa HS, El-Latif AAA (2019) Detection of Myocardial Infarction Based on Novel Deep Transfer Learning Methods for Urban Healthcare in Smart Cities. arXiv preprint arXiv:1906.09358
Alom MZ, Taha TM, Yakopcic C, Westberg S, Sidike P, Nasrin MS, Asari VK (2019) A state-of-the-art survey on deep learning theory and architectures. Electronics 8(3):292
Alquran H, Alqudah A, Abu-Qasmieh I, Al-Badarneh A, Almashaqbeh S (2019) ECG classification using higher order spectral estimation and deep learning techniques. Neural Netw World 29(4):207–219
Altan G, Allahverdi N, Kutlu Y (2018) A multistage deep learning algorithm for detecting arrhythmia. Paper presented at the 2018 1st international conference on computer applications & information security (ICCAIS)
Amrani M, Hammad M, Jiang F, Wang K, Amrani A (2018) Very deep feature extraction and fusion for arrhythmias detection. Neural Comput Appl 30(7):2047–2057
Andersen RS, Peimankar A, Puthusserypady S (2019) A deep learning approach for real-time detection of atrial fibrillation. Expert Syst Appl 115:465–473
Antczak K (2018) Deep recurrent neural networks for ECG signal denoising. arXiv preprint arXiv:1807.11551
Apandi ZFM, Ikeura R, Hayakawa S (2018) Arrhythmia Detection Using MIT-BIH Dataset: A Review. Paper presented at the 2018 International Conference on Computational Approach in Smart Systems Design and Applications (ICASSDA)
Assodiky H, Syarif I, Badriyah T (2017) Deep learning algorithm for arrhythmia detection. Paper presented at the 2017 International Electronics Symposium on Knowledge Creation and Intelligent Computing (IES-KCIC)
Azimi I, Takalo-Mattila J, Anzanpour A, Rahmani AM, Soininen J-P, Liljeberg P (2018) Empowering healthcare IoT systems with hierarchical edge-based deep learning. Paper presented at the Proceedings of the 2018 IEEE/ACM International Conference on Connected Health: Applications, Systems and Engineering Technologies
Babar MI, Ghazali M, Jawawi DN (2014) Systematic reviews in requirements engineering: A systematic review. Paper presented at the 2014 8th. Malaysian Software Engineering Conference (MySEC)
Bajare SR, Ingale VV (2019) ECG Based Biometric for Human Identification using Convolutional Neural Network. Paper presented at the 2019 10th International Conference on Computing, Communication and Networking Technologies (ICCCNT)
Baloglu UB, Talo M, Yildirim O, San Tan R, Acharya UR (2019) Classification of myocardial infarction with multi-lead ECG signals and deep CNN. Pattern Recognit Lett 122:23–30
Banerjee R, Ghose A, Khandelwal S (2019) A Novel Recurrent Neural Network Architecture for Classification of Atrial Fibrillation Using Single-lead ECG. Paper presented at the 2019 27th European Signal Processing Conference (EUSIPCO)
Banluesombatkul N, Rakthanmanon T, Wilaiprasitporn T (2018) Single channel ECG for obstructive sleep apnea severity detection using a deep learning approach. Paper presented at the TENCON 2018–2018 IEEE Region 10 Conference
Bansal R, Sehgal P, Bedi P (2011) Minutiae extraction from fingerprint images-a review. IJCSI Int J Comput Sci 8(5):74–85
Belle A, Thiagarajan R, Soroushmehr S, Navidi F, Beard DA, Najarian K (2015) Big data analytics in healthcare. BioMed research international, 2015
Bengio Y, Simard P, Frasconi P (1994) Learning long-term dependencies with gradient descent is difficult. IEEE Trans Neural Networks 5(2):157–166
Bhardwaj R, Natrajan P, Balasubramanian V (2018) Study to Determine the Effectiveness of Deep Learning Classifiers for ECG Based Driver Fatigue Classification. Paper presented at the 2018 IEEE 13th International Conference on Industrial and Information Systems (ICIIS)
Biel L, Pettersson O, Philipson L, Wide P (2001) ECG analysis: a new approach in human identification. IEEE Trans Instrum Meas 50(3):808–812
Bizopoulos P, Koutsouris D (2018) Deep learning in cardiology. IEEE Rev Biomed Eng 12:168–193
Bote-Curiel L, Munoz-Romero S, Gerrero-Curieses A, Rojo-Álvarez JL (2019) Deep learning and big data in healthcare: A double review for critical beginners. Appl Sci 9(11):2331
Bowyer KW, Hollingsworth K, Flynn PJ (2008) Image understanding for iris biometrics: A survey. Comput Vis Image Underst 110(2):281–307
Brereton P, Kitchenham BA, Budgen D, Turner M, Khalil M (2007) Lessons from applying the systematic literature review process within the software engineering domain. J Syst Softw 80(4):571–583
Brito C, Machado A, Sousa A (2019) Electrocardiogram Beat-Classification Based on a ResNet Network. Paper presented at the MedInfo
Buyya R, Srirama SN, Casale G, Calheiros R, Simmhan Y, Varghese B, Netto MA (2018) A manifesto for future generation cloud computing: Research directions for the next decade. ACM Comput Surv (CSUR) 51(5):1–38
Byeon Y-H, Kwak K-C (2019) Pre-configured deep convolutional neural networks with various time-frequency representations for biometrics from ECG signals. Appl Sci 9(22):4810
Byeon Y-H, Pan S-B, Kwak K-C Ensemble Deep Learning Models for ECG-based Biometrics. Paper presented at the 2020 Cybernetics & Informatics (K&I)
Byeon Y-H, Pan S-B, Kwak K-C (2019) Intelligent deep models based on scalograms of electrocardiogram signals for biometrics. Sensors 19(4):935
Byeon Y-H, Pan S-B, Kwak K-C (2020) Ensemble Deep Learning Models for ECG-based Biometrics. Paper presented at the 2020 Cybernetics & Informatics (K&I)
Cai W, Chen Y, Guo J, Han B, Shi Y, Ji L, Luo J (2020) Accurate detection of atrial fibrillation from 12-lead ECG using deep neural network. Comput Biol Med 116:103378
Carreiras C, Lourenço A, Silva H, Fred A, Ferreira R (2016) Evaluating template uniqueness in ECG biometrics. Informatics in Control, Automation and Robotics. Springer, pp 111–123
Chamatidis I, Katsika A, Spathoulas G (2017) Using deep learning neural networks for ECG based authentication. Paper presented at the 2017 International Carnahan Conference on Security Technology (ICCST)
Chang K-C, Hsieh P-H, Wu M-Y, Wang Y-C, Chen J-Y, Tsai F-J, Huang T-C (2020) Usefulness of Machine-Learning-Based Detection and Classification of Cardiac Arrhythmias with 12-Lead Electrocardiograms.Canadian Journal of Cardiology
Chen C, Hua Z, Zhang R, Liu G, Wen W (2020) Automated arrhythmia classification based on a combination network of CNN and LSTM. Biomed Signal Process Control 57:101819
Chen Y-J, Liu C-L, Tseng VS, Hu Y-F, Chen S-A (2019) Large-scale Classification of 12-lead ECG with Deep Learning. Paper presented at the 2019 IEEE EMBS International Conference on Biomedical & Health Informatics (BHI)
Chen Y, Chen H, He Z, Yang C, Cao Y (2018) Multi-Channel Lightweight Convolution Neural Network for Anterior Myocardial Infarction Detection. Paper presented at the 2018 IEEE SmartWorld, Ubiquitous Intelligence & Computing, Advanced & Trusted Computing, Scalable Computing & Communications, Cloud & Big Data Computing, Internet of People and Smart City Innovation (SmartWorld/SCALCOM/UIC/ATC/CBDCom/IOP/SCI)
Chen Y, Chen W (2018) Finger ECG based two-phase authentication using 1D convolutional neural networks. Paper presented at the 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC)
Cheng M, Sori WJ, Jiang F, Khan A, Liu S (2017) Recurrent neural network based classification of ECG signal features for obstruction of sleep apnea detection. Paper presented at the 2017 IEEE International Conference on Computational Science and Engineering (CSE) and IEEE International Conference on Embedded and Ubiquitous Computing (EUC)
Cho H-M, Park H, Dong S-Y, Youn I (2019) Ambulatory and Laboratory Stress Detection Based on Raw Electrocardiogram Signals Using a Convolutional Neural Network. Sensors 19(20):4408
Chollet F (2017) Xception: Deep learning with depthwise separable convolutions. Paper presented at the Proceedings of the IEEE conference on computer vision and pattern recognition
Chollet F (2018) Deep Learning mit Python und Keras: Das Praxis-Handbuch vom Entwickler der Keras-Bibliothek. MITP-Verlags GmbH & Co. KG
Chu J, Wang H, Lu W (2019) A novel two-lead arrhythmia classification system based on CNN and LSTM. J Mech Med Biology 19(03):1950004
Clifford GD, Liu C, Moody B, Li-wei HL, Silva I, Li Q, Mark RG (2017) AF Classification from a short single lead ECG recording: the PhysioNet/Computing in Cardiology Challenge 2017. Paper presented at the 2017 Computing in Cardiology (CinC)
Corradi F, Buil J, De Cannière H, Groenendaal W, Vandervoort P (2019) Real Time Electrocardiogram Annotation with a Long Short Term Memory Neural Network. Paper presented at the 2019 IEEE Biomedical Circuits and Systems Conference (BioCAS)
da Silva HP, Carreiras C, Lourenço A, Fred A, das Neves RC, Ferreira R (2015) Off-the-person electrocardiography: performance assessment and clinical correlation. Health and Technology 4(4):309–318
Dang H, Sun M, Zhang G, Qi X, Zhou X, Chang Q (2019) A novel deep arrhythmia-diagnosis network for atrial fibrillation classification using electrocardiogram signals. IEEE Access 7:75577–75590
Dang H, Sun M, Zhang G, Zhou X, Chang Q, Xu X (2019) A novel deep convolutional neural network for arrhythmia classification. Paper presented at the 2019 International Conference on Advanced Mechatronic Systems (ICAMechS)
Darmawahyuni A, Nurmaini S (2019) Deep Learning with Long Short-Term Memory for Enhancement Myocardial Infarction Classification. Paper presented at the 2019 6th International Conference on Instrumentation, Control, and Automation (ICA)
Darmawahyuni A, Nurmaini S, Caesarendra W, Bhayyu V, Rachmatullah MN (2019) Deep Learning with a Recurrent Network Structure in the Sequence Modeling of Imbalanced Data for ECG-Rhythm Classifier. Algorithms 12(6):118
Debnath T, Biswas T, Ashik MH, Dash S (2018) Auto-encoder based nonlinear dimensionality reduction of ecg data and classification of cardiac arrhythmia groups using deep neural network. Paper presented at the 2018 4th International Conference on Electrical Engineering and Information & Communication Technology (iCEEiCT)
Deshmane M, Madhe S (2018) ECG based biometric human identification using convolutional neural network in smart health applications. Paper presented at the 2018 Fourth International Conference on Computing Communication Control and Automation (ICCUBEA)
Dey D, Chaudhuri S, Munshi S (2018) Obstructive sleep apnoea detection using convolutional neural network based deep learning framework. Biomed Eng Lett 8(1):95–100
Diker A, Engin A (2019) Feature extraction of ECG signal by using deep feature. Paper presented at the 2019 7th International Symposium on Digital Forensics and Security (ISDFS)
Ding Z, Qiu S, Guo Y, Lin J, Sun L, Fu D, Meng L (2019) LabelECG: A Web-Based Tool for Distributed Electrocardiogram Annotation Machine Learning and Medical Engineering for Cardiovascular Health and Intravascular Imaging and Computer Assisted Stenting (pp. 104–111): Springer
Dokur Z, Ölmez T (2020) Heartbeat classification by using a convolutional neural network trained with Walsh functions.Neural Computing and Applications,1–20
Eduardo A, Aidos H, Fred A (2017) ECG-based biometrics using a deep autoencoder for feature learning-an empirical study on transferability. Paper presented at the International Conference on Pattern Recognition Applications and Methods
Emmert-Streib F, Yang Z, Feng H, Tripathi S, Dehmer M (2020) An Introductory Review of Deep Learning for Prediction Models With Big Data. Front.Artif. Intell, 3(4)
Erdenebayar U, Kim H, Park J-U, Kang D, Lee K-J (2019) Automatic prediction of atrial fibrillation based on convolutional neural network using a short-term normal electrocardiogram signal. Journal of Korean medical science, 34(7)
Erdenebayar U, Kim YJ, Park J-U, Joo EY, Lee K-J (2019) Deep learning approaches for automatic detection of sleep apnea events from an electrocardiogram. Comput Methods Programs Biomed 180:105001
Farahani B, Barzegari M, Aliee FS (2019) Towards collaborative machine learning driven healthcare internet of things. Paper presented at the Proceedings of the International Conference on Omni-Layer Intelligent Systems
Farhadi J, Attarodi G, Dabanloo NJ, Mohandespoor M, Eslamizadeh M (2018) Classification of atrial fibrillation using stacked auto encoders neural networks. Paper presented at the 2018 Computing in Cardiology Conference (CinC)
Faust O, Hagiwara Y, Hong TJ, Lih OS, Acharya UR (2018) Deep learning for healthcare applications based on physiological signals: A review. Comput Methods Programs Biomed 161:1–13
Faust O, Shenfield A, Kareem M, San TR, Fujita H, Acharya UR (2018) Automated detection of atrial fibrillation using long short-term memory network with RR interval signals. Comput Biol Med 102:327–335
Fischer A, Igel C (2012) An introduction to restricted Boltzmann machines. Paper presented at the Iberoamerican congress on pattern recognition
Fischer A, Igel C (2014) Training restricted Boltzmann machines: An introduction. Pattern Recogn 47(1):25–39
Forsen G, Nelson M, Staron R Jr (1977) Personal Attributes Authentication Techniques; Pattern Analysis & Recognition Corporation, Rome Air Development Center. St. Utica, NY, USA
Fujita H, Cimr D (2019) Decision support system for arrhythmia prediction using convolutional neural network structure without preprocessing. Appl Intell 49(9):3383–3391
Fukushima K, Shiotani et al (1995) Neurocomputing, 9
Ganapathy N, Swaminathan R, Deserno TM (2018) Deep learning on 1-D biosignals: a taxonomy-based survey. Yearb Med Inform 27(01):098–109
Ghaffari A, Madani N (2019) Atrial fibrillation identification based on a deep transfer learning approach. Biomedical Phys Eng Express 5(3):035015
Giannakakis G, Trivizakis E, Tsiknakis M, Marias K (2019) A novel multi-kernel 1D convolutional neural network for stress recognition from ECG. Paper presented at the 2019 8th International Conference on Affective Computing and Intelligent Interaction Workshops and Demos (ACIIW)
Goodfellow I, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S, Bengio Y (2014) Generative adversarial nets. Paper presented at the Advances in neural information processing systems
Granados J, Chu H, Zou Z, Zheng L-R (2019) Towards Workload-Balanced, Live Deep Learning Analytics for Confidentiality-Aware IoT Medical Platforms. Paper presented at the 2019 IEEE International Conference on Artificial Intelligence Circuits and Systems (AICAS)
Guo L, Sim G, Matuszewski B (2019) Inter-patient ECG classification with convolutional and recurrent neural networks. Biocybernetics and Biomedical Engineering 39(3):868–879
Guo T, Fan Y, Chen M, Wu X, Zhang L, He T, Lu Z (2020) Cardiovascular implications of fatal outcomes of patients with coronavirus disease 2019 (COVID-19). JAMA Cardiol 5(7):811–818
Hafemann LG, Sabourin R, Oliveira LS (2017) Offline handwritten signature verification—Literature review. Paper presented at the 2017 Seventh International Conference on Image Processing Theory, Tools and Applications (IPTA)
Hammad M, Liu Y, Wang K (2018) Multimodal biometric authentication systems using convolution neural network based on different level fusion of ECG and fingerprint. IEEE Access 7:26527–26542
Hammad M, Pławiak P, Wang K, Acharya UR (2020) ResNet-Attention model for human authentication using ECG signals.Expert Systems, e12547
Hammad M, Wang K (2019) Parallel score fusion of ECG and fingerprint for human authentication based on convolution neural network. Computers & Security 81:107–122
Hammad M, Zhang S, Wang K (2019) A novel two-dimensional ECG feature extraction and classification algorithm based on convolution neural network for human authentication. Future Generation Computer Systems 101:180–196
Han S-Y, Kwak N-S, Oh T, Lee S-W (2020) Classification of pilots’ mental states using a multimodal deep learning network. Biocybernetics and Biomedical Engineering 40(1):324–336
Hao C, Wibowo S, Majmudar M, Rajput KS (2019) Spectro-Temporal Feature Based Multi-Channel Convolutional Neural Network for ECG Beat Classification. Paper presented at the 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC)
Hartmann M, Farooq H, Imran A (2019) Distilled Deep Learning based Classification of Abnormal Heartbeat Using ECG Data through a Low Cost Edge Device. Paper presented at the 2019 IEEE Symposium on Computers and Communications (ISCC)
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. Paper presented at the Proceedings of the IEEE conference on computer vision and pattern recognition
Hernández-Blanco, A., Herrera-Flores, B., Tomás, D., & Navarro-Colorado, B. (2019). A systematic review of deep learning approaches to educational data mining. Complexity: https://doi.org/10.1155/2019/1306039
Hinton GE, Osindero S, Teh Y-W (2006) A fast learning algorithm for deep belief nets. Neural Comput 18(7):1527–1554
Hinton GE, Salakhutdinov RR (2006) Reducing the dimensionality of data with neural networks. Science 313(5786):504–507
Hochreiter S, Schmidhuber J (1997) Long short-term memory. Neural Comput 9(8):1735–1780
Hong P-L, Hsiao J-Y, Chung C-H, Feng Y-M, Wu S-C (2019) ECG biometric recognition: template-free approaches based on deep learning. Paper presented at the 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC)
Hong S, Zhou Y, Shang J, Xiao C, Sun J (2019) Opportunities and Challenges in Deep Learning Methods on Electrocardiogram Data: A Systematic Review. arXiv arXiv preprint arXiv:2001.01550
Hong S, Zhou Y, Shang J, Xiao C, Sun J (2020) Opportunities and challenges of deep learning methods for electrocardiogram data: A systematic review.Computers in biology and medicine,103801
Hsieh C-H, Li Y-S, Hwang B-J, Hsiao C-H (2020) Detection of Atrial Fibrillation Using 1D Convolutional Neural Network. Sensors 20(7):2136
Huang G, Liu Z, Van Der Maaten L, Weinberger KQ (2017) Densely connected convolutional networks. Paper presented at the Proceedings of the IEEE conference on computer vision and pattern recognition
Huang J, Chen B, Yao B, He W (2019) ECG arrhythmia classification using STFT-based spectrogram and convolutional neural network. IEEE Access 7:92871–92880
Huang M-L, Wu Y-S (2020) Classification of atrial fibrillation and normal sinus rhythm based on convolutional neural network.Biomedical engineering letters,1–11
Huanhuan M, Yue Z (2014) Classification of electrocardiogram signals with deep belief networks. Paper presented at the 2014 IEEE 17th International Conference on Computational Science and Engineering
Hubel DH, Wiesel TN (1962) Receptive fields, binocular interaction and functional architecture in the cat’s visual cortex. J Physiol 160(1):106
Iandola FN, Han S, Moskewicz MW, Ashraf K, Dally WJ, Keutzer K (2016) SqueezeNet: AlexNet-level accuracy with 50x fewer parameters and < 0.5 MB model size. arXiv preprint arXiv:1602.07360
Ince T, Zabihi M, Kiranyaz S, Gabbouj M (2017) Learned vs. Hand-Designed Features for ECG Beat Classification: A Comprehensive. In: Study (ed) EMBEC & NBC 2017. Springer, pp 551–554
Irvine JM, Israel SA, Wiederhold MD, Wiederhold BK (2003) A new biometric: human identification from circulatory function. Paper presented at the Joint Statistical Meetings of the American Statistical Association, San Francisco
Isin A, Ozdalili S (2017) Cardiac arrhythmia detection using deep learning. Procedia Comput Sci 120:268–275
Ivanovic MD, Atanasoski V, Shvilkin A, Hadzievski L, Maluckov A (2019) Deep Learning Approach for Highly Specific Atrial Fibrillation and Flutter Detection based on RR Intervals. Paper presented at the 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC)
Izci E, Ozdemir MA, Degirmenci M, Akan A (2019) Cardiac Arrhythmia Detection from 2D ECG Images by Using Deep Learning Technique. Paper presented at the 2019 Medical Technologies Congress (TIPTEKNO)
Jabbar R, Al-Khalifa K, Kharbeche M, Alhajyaseen W, Jafari M, Jiang S (2018) Real-time driver drowsiness detection for android application using deep neural networks techniques. Procedia Comput Sci 130:400–407
Jeon E, Chae M, Han S, Lee H (2019) Arrhythmia Classification System Using Deep Neural Network. Paper presented at the 2019 Eleventh International Conference on Ubiquitous and Future Networks (ICUFN)
Jiang F, Jiang Y, Zhi H, Dong Y, Li H, Ma S, Wang Y (2017) Artificial intelligence in healthcare: past, present and future. Stroke and vascular neurology 2(4):230–243
Jun TJ, Park HJ, Minh NH, Kim D, Kim Y-H (2016) Premature ventricular contraction beat detection with deep neural networks. Paper presented at the 2016 15th IEEE International Conference on Machine Learning and Applications (ICMLA)
Kachuee M, Fazeli S, Sarrafzadeh M (2018) Ecg heartbeat classification: A deep transferable representation. Paper presented at the 2018 IEEE International Conference on Healthcare Informatics (ICHI)
Kamaleswaran R, Mahajan R, Akbilgic O (2018) A robust deep convolutional neural network for the classification of abnormal cardiac rhythm using single lead electrocardiograms of variable length. Physiol Meas 39(3):035006
Kaouter K, Mohamed T, Sofiene D, Abbas D, Fouad M (2019) Full training convolutional neural network for ECG signals classification. Paper presented at the AIP Conference Proceedings
Kayalibay B, Jensen G, van der Smagt P (2017) CNN-based segmentation of medical imaging data. arXiv preprint arXiv:1701.03056
Khan S, Rahmani H, Shah SAA, Bennamoun M (2018) A guide to convolutional neural networks for computer vision. Synthesis Lectures on Computer Vision 8(1):1–207
Kim JS, Kim SH, Pan SB (2020) Personal recognition using convolutional neural network with ECG coupling image. J Ambient Intell Humaniz Comput 11(5):1923–1932
Kim KG (2016) Book review: Deep learning. Healthc Inf Res 22(4):351–354
Kim M-G, Choi C, Pan SB (2020) Ensemble Networks for User Recognition in Various Situations Based on Electrocardiogram. IEEE Access 8:36527–36535
Kiranyaz S, Ince T, Gabbouj M (2015) Real-time patient-specific ECG classification by 1-D convolutional neural networks. IEEE Trans Biomed Eng 63(3):664–675
Kitchenham B, Charters S (2007) Guidelines for performing systematic literature reviews in software engineering version 2.3. Engineering 45(4ve):1051
Konan DAM, Patel W (2018) i-NXGeVita: IoMT based Ubiquitous Health Monitoring System using Deep Neural Networks. Paper presented at the 2018 3rd IEEE International Conference on Recent Trends in Electronics, Information & Communication Technology (RTEICT)
Krizhevsky A, Sutskever I, Hinton GE (2012) Imagenet classification with deep convolutional neural networks. Paper presented at the Advances in neural information processing systems
Kulik S, Shtanko A (2020) Using convolutional neural networks for recognition of objects varied in appearance in computer vision for intellectual robots. Procedia Comput Sci 169:164–167
Kusuma S, Udayan JD (2020) Analysis on Deep Learning methods for ECG based Cardiovascular Disease prediction. Scalable Computing: Practice and Experience 21(1):127–136
Kwon J-M, Jeon K-H, Kim HM, Kim MJ, Lim SM, Kim K-H, Oh B-H (2020) Comparing the performance of artificial intelligence and conventional diagnosis criteria for detecting left ventricular hypertrophy using electrocardiography. EP Europace 22(3):412–419
Labati RD, Muñoz E, Piuri V, Sassi R, Scotti F (2019) Deep-ECG: convolutional neural networks for ECG biometric recognition. Pattern Recognit Lett 126:78–85
LeCun Y, Bottou L, Bengio Y, Haffner P (1998) Gradient-based learning applied to document recognition. Proceedings of the IEEE, 86(11), 2278–2324
Lee J-G, Jun S, Cho Y-W, Lee H, Kim GB, Seo JB, Kim N (2017) Deep learning in medical imaging: general overview. Korean J Radiol 18(4):570–584
Lei X, Zhang Y, Lu Z (2016) Deep learning feature representation for electrocardiogram identification. Paper presented at the 2016 IEEE International Conference on Digital Signal Processing (DSP)
Li D, Li X, Zhao J, Bai X (2019) Automatic staging model of heart failure based on deep learning. Biomed Signal Process Control 52:77–83
Li D, Zhang J, Zhang Q, Wei X (2017) Classification of ECG signals based on 1D convolution neural network. Paper presented at the 2017 IEEE 19th International Conference on e-Health Networking, Applications and Services (Healthcom)
Li J, Si Y, Xu T, Jiang S (2018) Deep convolutional neural network based ECG classification system using information fusion and one-hot encoding techniques. Mathematical Problems in Engineering, 2018
Li Y, Pang Y, Wang K, Li X (2020) Toward improving ECG biometric identification using cascaded convolutional neural networks. Neurocomputing
Li Z, Feng X, Wu Z, Yang C, Bai B, Yang Q (2019) Classification of atrial fibrillation recurrence based on a convolution neural network with SVM architecture. IEEE Access 7:77849–77856
Li Z, Yang C, Zhang Q (2018) Noninvasive Prediction of Atrial Fibrillation Recurrence Based on a Deep Learning Algorithm. Paper presented at the Proceedings of the 2018 2nd International Conference on Computational Biology and Bioinformatics
Li Z, Zhou D, Wan L, Li J, Mou W (2020) Heartbeat classification using deep residual convolutional neural network from 2-lead electrocardiogram. J Electrocardiol 58:105–112
Lih OS, Jahmunah V, San TR, Ciaccio EJ, Yamakawa T, Tanabe M, Acharya UR (2020) Comprehensive electrocardiographic diagnosis based on deep learning. Artif Intell Med 103:101789
Liu F, Liu C, Zhao L, Zhang X, Wu X, Xu X, He Z (2018) An open access database for evaluating the algorithms of electrocardiogram rhythm and morphology abnormality detection. J Med Imaging Health Inf 8(7):1368–1373
Liu W, Huang Q, Chang S, Wang H, He J (2018) Multiple-feature-branch convolutional neural network for myocardial infarction diagnosis using electrocardiogram. Biomed Signal Process Control 45:22–32
Lo F-W, Tsai P-Y (2018) Deep learning for detection of fetal ECG from multi-channel abdominal leads. Paper presented at the 2018 Asia-Pacific Signal and Information Processing Association Annual Summit and Conference (APSIPA ASC)
Lui HW, Chow KL (2018) Multiclass classification of myocardial infarction with convolutional and recurrent neural networks for portable ECG devices. Inf Med Unlocked 13:26–33
Luo K, Li J, Wang Z, Cuschieri A (2017) Patient-specific deep architectural model for ECG classification. Journal of healthcare engineering, 2017
Lynn HM, Pan SB, Kim P (2019) A deep bidirectional GRU network model for biometric electrocardiogram classification based on recurrent neural networks. IEEE Access 7:145395–145405
Ma J, Liu S, Chen G (2018) Identification and Classification of Electrocardiogram Signals Based on Convolutional Recurrent Neural Network. Paper presented at the 2018 11th International Congress on Image and Signal Processing, BioMedical Engineering and Informatics (CISP-BMEI)
Mathews SM, Kambhamettu C, Barner KE (2018) A novel application of deep learning for single-lead ECG classification. Comput Biol Med 99:53–62
McBee MP, Awan OA, Colucci AT, Ghobadi CW, Kadom N, Kansagra AP, Auffermann WF (2018) Deep learning in radiology. Acad Radiol 25(11):1472–1480
Mincholé A, Camps J, Lyon A, Rodríguez B (2019) Machine learning in the electrocardiogram. J Electrocardiol 57:S61–S64
Miotto R, Wang F, Wang S, Jiang X, Dudley JT (2018) Deep learning for healthcare: review, opportunities and challenges. Brief Bioinform 19(6):1236–1246
Mohammed M, Khan MB, Bashier EBM (2016) Machine learning: algorithms and applications. Crc Press
Monrose F, Rubin AD (2000) Keystroke dynamics as a biometric for authentication. Future Generation Computer Systems 16(4):351–359
Moody GB, Mark RG, Goldberger AL (2001) PhysioNet: a web-based resource for the study of physiologic signals. IEEE Eng Med Biol Mag 20(3):70–75
Mostafa SS, Mendonça F, Morgado-Dias F, Ravelo-García A (2017) SpO2 based sleep apnea detection using deep learning. Paper presented at the 2017 IEEE 21st international conference on intelligent engineering systems (INES)
Muhammed MA, Aravinth J (2019) CNN based Off-the-Person ECG Biometrics. Paper presented at the 2019 International Conference on Wireless Communications Signal Processing and Networking (WiSPNET)
Murugesan B, Ravichandran V, Ram K, Preejith S, Joseph J, Shankaranarayana SM, Sivaprakasam M (2018) Ecgnet: Deep network for arrhythmia classification. Paper presented at the 2018 IEEE International Symposium on Medical Measurements and Applications (MeMeA)
Nassif AB, Shahin I, Attili I, Azzeh M, Shaalan K (2019) Speech recognition using deep neural networks: A systematic review. IEEE Access 7:19143–19165
Nguyen G, Dlugolinsky S, Bobák M, Tran V, García ÁL, Heredia I, Hluchý L (2019) Machine Learning and Deep Learning frameworks and libraries for large-scale data mining: a survey. Artif Intell Rev 52(1):77–124
Oh SL, Ng EY, Tan S, Acharya UR (2018) Automated diagnosis of arrhythmia using combination of CNN and LSTM techniques with variable length heart beats. Comput Biol Med 102:278–287
Ouchicha C, Ammor O, Meknassi M (2020) CVDNet: A novel deep learning architecture for detection of coronavirus (Covid-19) from chest x-ray images. Chaos Solitons Fractals 140:110245
Page A, Kulkarni A, Mohsenin T (2015) Utilizing deep neural nets for an embedded ECG-based biometric authentication system. Paper presented at the 2015 IEEE Biomedical Circuits and Systems Conference (BioCAS)
Pak M, Kim S (2017) A review of deep learning in image recognition. Paper presented at the 2017 4th international conference on computer applications and information processing technology (CAIPT)
Pan SJ, Yang Q (2009) A survey on transfer learning. IEEE Trans Knowl Data Eng 22(10):1345–1359
Pandey SK, Janghel RR (2019) Automatic detection of arrhythmia from imbalanced ECG database using CNN model with SMOTE. Australasian Phys Eng Sci Med 42(4):1129–1139
Pandey SK, Janghel RR (2020) Automatic arrhythmia recognition from electrocardiogram signals using different feature methods with long short-term memory network model.Signal, Image and Video Processing,1–9
Park JY, Cho H, Hwang W, Balan RK, Ko JG (2019) Deep ECG estimation using a bed-attached geophone. Paper presented at the 17th ACM International Conference on Mobile Systems, Applications, and Services, MobiSys 2019
Picon A, Irusta U, Álvarez-Gila A, Aramendi E, Alonso-Atienza F, Figuera C, Kramer-Johansen J (2019) Mixed convolutional and long short-term memory network for the detection of lethal ventricular arrhythmia. PLoS ONE 14(5):e0216756
Pierson HA, Gashler MS (2017) Deep learning in robotics: a review of recent research. Adv Robot 31(16):821–835
Pinto JR, Cardoso JS, Lourenço A (2018) Evolution, current challenges, and future possibilities in ECG biometrics. IEEE Access 6:34746–34776
Polat Ç, Karaman O, Karaman C, Korkmaz G, Balcí MC, Kelek SE COVID-19 diagnosis from chest X-ray images using transfer learning: Enhanced performance by debiasing dataloader.Journal of X-Ray Science and Technology(Preprint),1–18
Pourbabaee B, Roshtkhari MJ, Khorasani K (2018) Deep convolutional neural networks and learning ECG features for screening paroxysmal atrial fibrillation patients. IEEE Trans Syst Man Cybernetics: Syst 48(12):2095–2104
Pyakillya B, Kazachenko N, Mikhailovsky N (2017) Deep learning for ECG classification. Paper presented at the Journal of physics: conference series
Qayyum A, Meriaudeau F, Chan GC (2018) Classification of atrial fibrillation with pre-trained convolutional neural network models. Paper presented at the 2018 IEEE-EMBS Conference on Biomedical Engineering and Sciences (IECBES)
Qayyum ABA, Islam T, Haque MA (2019) ECG Heartbeat Classification: A Comparative Performance Analysis between One and Two Dimensional Convolutional Neural Network. Paper presented at the 2019 IEEE International Conference on Biomedical Engineering, Computer and Information Technology for Health (BECITHCON)
Rajkumar A, Ganesan M, Lavanya R (2019) Arrhythmia classification on ECG using Deep Learning. Paper presented at the 2019 5th International Conference on Advanced Computing & Communication Systems (ICACCS)
Ranjan A (2019) Permanence of ECG Biometric: Experiments Using Convolutional Neural Networks. Paper presented at the 2019 International Conference on Biometrics (ICB)
Rastgoo MN, Nakisa B, Maire F, Rakotonirainy A, Chandran V (2019) Automatic driver stress level classification using multimodal deep learning. Expert Syst Appl 138:112793
Ribeiro AH, Ribeiro MH, Paixão GM, Oliveira DM, Gomes PR, Canazart JA, Wagner M Jr (2020) Automatic diagnosis of the 12-lead ECG using a deep neural network. Nat Commun 11(1):1–9
Ribeiro ALP, Paixão GM, Gomes PR, Ribeiro MH, Ribeiro AH, Canazart JA, de Moraes JL (2019) Tele-electrocardiography and bigdata: The CODE (Clinical Outcomes in Digital Electrocardiography) study. J Electrocardiol 57:S75–S78
Rim B, Sung N-J, Min S, Hong M (2020) Deep Learning in Physiological Signal Data: A Survey. Sensors 20(4):969
Rubin J, Parvaneh S, Rahman A, Conroy B, Babaeizadeh S (2018) Densely connected convolutional networks for detection of atrial fibrillation from short single-lead ECG recordings. J Electrocardiol 51(6):S18–S21
Saadatnejad S, Oveisi M, Hashemi M (2019) LSTM-based ECG classification for continuous monitoring on personal wearable devices. IEEE J biomedical health Inf 24(2):515–523
Salloum R, Kuo C-CJ (2017) ECG-based biometrics using recurrent neural networks. Paper presented at the 2017 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP)
Sannino G, De Pietro G (2018) A deep learning approach for ECG-based heartbeat classification for arrhythmia detection. Future Generation Computer Systems 86:446–455
Sarkar I, Alisherov F, Kim T-h, Bhattacharyya D (2010) Palm vein authentication system: a review.International Journal of Control and Automation, 3(1)
Schmidhuber J (2015) Deep learning in neural networks: An overview. Neural Netw 61:85–117
Sengupta S, Basak S, Saikia P, Paul S, Tsalavoutis V, Atiah F, Peters A (2020) A review of deep learning with special emphasis on architectures, applications and recent trends.Knowledge-Based Systems,105596
Serhani MA, El Kassabi T, Ismail H, Navaz N, A (2020) ECG Monitoring Systems: Review, Architecture, Processes, and Key Challenges. Sensors 20(6):1796
Shaker AM, Tantawi M, Shedeed HA, Tolba MF (2020) Generalization of Convolutional Neural Networks for ECG Classification Using Generative Adversarial Networks. IEEE Access 8:35592–35605
Shankar MG, Babu CG (2020) An exploration of ECG signal feature selection and classification using machine learning techniques. Int. J. Innovative Technol. Exploring Eng. Regul(2020), 9
Sharma A, Garg N, Patidar S, San Tan R, Acharya UR (2020) Automated pre-screening of arrhythmia using hybrid combination of Fourier-Bessel expansion and LSTM. Computers in biology and medicine, 103753
Shashikumar SP, Shah AJ, Clifford GD, Nemati S (2018) Detection of paroxysmal atrial fibrillation using attention-based bidirectional recurrent neural networks. Paper presented at the Proceedings of the 24th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining
shensheng Xu S, Mak M-W, Cheung C-C (2017) Deep neural networks versus support vector machines for ECG arrhythmia classification. Paper presented at the 2017 IEEE International Conference on Multimedia & Expo Workshops (ICMEW)
Shepley AJ (2019) Deep Learning For Face Recognition: A Critical Analysis. arXiv preprint arXiv:1907.12739
Shi H, Qin C, Xiao D, Zhao L, Liu C (2020) Automated heartbeat classification based on deep neural network with multiple input layers. Knowl Based Syst 188:105036
Shi H, Wang H, Qin C, Zhao L, Liu C (2020) An incremental learning system for atrial fibrillation detection based on transfer learning and active learning. Comput Methods Programs Biomed 187:105219
Shrestha A, Mahmood A (2019) Review of deep learning algorithms and architectures. IEEE Access 7:53040–53065
Simonyan K, Zisserman A (2014) Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556
Singh S, Pandey SK, Pawar U, Janghel RR (2018) Classification of ECG arrhythmia using recurrent neural networks. Procedia Comput Sci 132:1290–1297
Smolensky P (1986) Information processing in dynamical systems: Foundations of harmony theory. Colorado Univ at Boulder Dept of Computer Science
Sugimoto K, Lee S, Okada Y (2018) Deep learning-based detection of periodic abnormal waves in ecg data. Paper presented at the Proceedings of the International MultiConference of Engineers and Computer Scientists
Sujadevi V, Soman K, Vinayakumar R (2017) Real-time detection of atrial fibrillation from short time single lead ECG traces using recurrent neural networks. Paper presented at the The International Symposium on Intelligent Systems Technologies and Applications
Swapna G, Kp S, Vinayakumar R (2018) Automated detection of diabetes using CNN and CNN-LSTM network and heart rate signals. Procedia Comput Sci 132:1253–1262
Swapna G, Soman K, Vinayakumar R (2018) Automated detection of cardiac arrhythmia using deep learning techniques. Procedia Comput Sci 132:1192–1201
Swapna G, Vinayakumar R, Soman K (2018) Diabetes detection using deep learning algorithms. ICT Express 4(4):243–246
Szegedy C, Liu W, Jia Y, Sermanet P, Reed S, Anguelov D, Rabinovich A (2015) Going deeper with convolutions. Paper presented at the Proceedings of the IEEE conference on computer vision and pattern recognition
Taherisadr M, Asnani P, Galster S, Dehzangi O (2018) ECG-based driver inattention identification during naturalistic driving using Mel-frequency cepstrum 2-D transform and convolutional neural networks. Smart Health 9:50–61
Takalo-Mattila J, Kiljander J, Soininen J-P (2018) Inter-patient ECG classification using deep convolutional neural networks. Paper presented at the 2018 21st Euromicro Conference on Digital System Design (DSD)
Tan JH, Hagiwara Y, Pang W, Lim I, Oh SL, Adam M, Acharya UR (2018) Application of stacked convolutional and long short-term memory network for accurate identification of CAD ECG signals. Comput Biol Med 94:19–26
Tobore I, Li J, Yuhang L, Al-Handarish Y, Kandwal A, Nie Z, Wang L (2019) Deep learning intervention for health care challenges: some biomedical domain considerations.JMIR mHealth and uHealth, 7(8), e11966
Upadhya V, Sastry P (2019) An overview of restricted Boltzmann machines.Journal of the Indian Institute of Science,1–12
Urtnasan E, Park J-U, Joo E-Y, Lee K-J (2018) Automated detection of obstructive sleep apnea events from a single-lead electrocardiogram using a convolutional neural network. J Med Syst 42(6):104
Urtnasan E, Park J-U, Lee K-J (2018) Multiclass classification of obstructive sleep apnea/hypopnea based on a convolutional neural network from a single-lead electrocardiogram. Physiol Meas 39(6):065003
Varghese B, Buyya R (2018) Next generation cloud computing: New trends and research directions. Future Generation Computer Systems 79:849–861
Verma D, Agarwal S (2018) Cardiac Arrhythmia Detection from Single-lead ECG using CNN and LSTM assisted by Oversampling. Paper presented at the 2018 International Conference on Advances in Computing, Communications and Informatics (ICACCI)
Voulodimos A, Doulamis N, Doulamis A, Protopapadakis E (2018) Deep learning for computer vision: A brief review. Computational intelligence and neuroscience, 2018
Vullings R (2019) Fetal Electrocardiography and Deep Learning for Prenatal Detection of Congenital Heart Disease. Paper presented at the 2019 Computing in Cardiology (CinC)
Wang EK, Xi L, Sun RP, Wang F, Pan L, Cheng C, Li Y (2019) A new deep learning model for assisted diagnosis on electrocardiogram. Math Biosci engineering: MBE 16(4):2481–2491
Wang X (2016) Deep learning in object recognition, detection, and segmentation. Found Trends Signal Process 8(4):217–382
Wang Y, Liao W, Chang Y (2018) Gated recurrent unit network-based short-term photovoltaic forecasting. Energies 11(8):2163
Weiss K, Khoshgoftaar TM, Wang D (2016) A survey of transfer learning. J Big data 3(1):9
Wieclaw L, Khoma Y, Fałat P, Sabodashko D, Herasymenko V (2017) Biometrie identification from raw ECG signal using deep learning techniques. Paper presented at the 2017 9th IEEE International Conference on Intelligent Data Acquisition and Advanced Computing Systems: Technology and Applications (IDAACS)
Wu Z, Ding X, Zhang G (2016) A novel method for classification of ECG arrhythmias using deep belief networks. Int J Comput Intell Appl 15(04):1650021
Xi P, Law A, Goubran R, Shu C (2019) Pilot Workload Prediction from ECG Using Deep Convolutional Neural Networks. Paper presented at the 2019 IEEE International Symposium on Medical Measurements and Applications (MeMeA)
Xia Y, Wulan N, Wang K, Zhang H (2018) Detecting atrial fibrillation by deep convolutional neural networks. Comput Biol Med 93:84–92
Xiao Y, Watson M (2019) Guidance on conducting a systematic literature review. J Plann Educ Res 39(1):93–112
Xiong Z, Stiles MK, Zhao J (2017) Robust ECG signal classification for detection of atrial fibrillation using a novel neural network. Paper presented at the 2017 Computing in Cardiology (CinC)
Xu SS, Mak M-W, Cheung C-C (2018) Towards end-to-end ECG classification with raw signal extraction and deep neural networks. IEEE J biomedical health Inf 23(4):1574–1584
Xu X, Liu H (2020) ECG heartbeat classification using convolutional neural networks. IEEE Access 8:8614–8619
Yamashita R, Nishio M, Do RKG, Togashi K (2018) Convolutional neural networks: an overview and application in radiology. Insights into imaging 9(4):611–629
Yang H-C, Islam MM, Li Y-C (2018) Potentiality of deep learning application in healthcare. Comput Methods Programs Biomed 161:1
Yao Z-J, Bi J, Chen Y-X (2018) Applying deep learning to individual and community health monitoring data: A survey. Int J Autom Comput 15(6):643–655
Yao Z, Chen Y (2018) Arrhythmia classification from single lead ecg by multi-scale convolutional neural networks. Paper presented at the Conf Proc IEEE Eng Med Biol Soc
Yildirim Ö (2018) A novel wavelet sequence based on deep bidirectional LSTM network model for ECG signal classification. Comput Biol Med 96:189–202
Yildirim O, Baloglu UB, Tan R-S, Ciaccio EJ, Acharya UR (2019) A new approach for arrhythmia classification using deep coded features and LSTM networks. Comput Methods Programs Biomed 176:121–133
Yıldırım Ö, Pławiak P, Tan R-S, Acharya UR (2018) Arrhythmia detection using deep convolutional neural network with long duration ECG signals. Comput Biol Med 102:411–420
Yin W, Kann K, Yu M, Schütze H (2017) Comparative study of cnn and rnn for natural language processing. arXiv preprint arXiv:1702.01923
Young T, Hazarika D, Poria S, Cambria E (2018) Recent trends in deep learning based natural language processing. IEEE Comput Intell Mag 13(3):55–75
Yuan C, Yan Y, Zhou L, Bai J, Wang L (2016) Automated atrial fibrillation detection based on deep learning network. Paper presented at the 2016 IEEE International Conference on Information and Automation (ICIA)
Zeiler MD, Fergus R (2014) Visualizing and understanding convolutional networks. Paper presented at the European conference on computer vision
Zhai J, Zhang S, Chen J, He Q (2018) Autoencoder and its various variants. Paper presented at the 2018 IEEE International Conference on Systems, Man, and Cybernetics (SMC)
Zhang D (2006) Wavelet approach for ECG baseline wander correction and noise reduction. Paper presented at the 2005 IEEE Engineering in Medicine and Biology 27th Annual Conference
Zhang Q, Zhou D (2018) Deep arm/ear-ECG image learning for highly wearable biometric human identification. Ann Biomed Eng 46(1):122–134
Zhang Q, Zhou D, Zeng X (2017) PulsePrint: Single-arm-ECG biometric human identification using deep learning. Paper presented at the 2017 IEEE 8th Annual Ubiquitous Computing, Electronics and Mobile Communication Conference (UEMCON)
Zhang Y, Yu J, Zhang Y, Liu C, Li J (2018) A convolutional neural network for identifying premature ventricular contraction beat and right bundle branch block beat. Paper presented at the 2018 International Conference on Sensor Networks and Signal Processing (SNSP)
Zhou S, Tan B (2020) Electrocardiogram soft computing using hybrid deep learning CNN-ELM. Appl Soft Comput 86:105778
Zhou Z, Zhai X, Tin C (2020) Fully Automatic Electrocardiogram Classification System based on Generative Adversarial Network with Auxiliary Classifier. arXiv preprint arXiv:2004.04894
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethics approval and consent to participate
All data originated from published articles.
Consent for publication
No individual person’s data was presented.
Conflict of interest
No competing interests are reported by the authors.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Musa, N., Gital, A.Y., Aljojo, N. et al. A systematic review and Meta-data analysis on the applications of Deep Learning in Electrocardiogram. J Ambient Intell Human Comput 14, 9677–9750 (2023). https://doi.org/10.1007/s12652-022-03868-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12652-022-03868-z